Quantitative Biology in Space and Time

Molecular Cell Biology
Head: Prof. Ann Ehrenhofer-Murray

Plant Physiology
Head: Prof. Bernhard Grimm

Optobiology
Head: Prof. Marina Mikhaylova


Structural Biology and Biochemistry
Head: Prof. Holger Dobbek

Plant Cell and Molecular Biology
Head: Prof. Kerstin Kaufmann
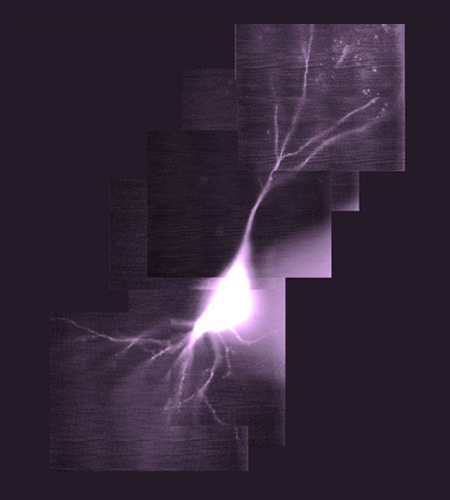

Cellular Biophysics
Head: Prof. Andrew Plested
In the Cellular Biophysics group, we study glutamate receptors and other components of fast synaptic transmission.
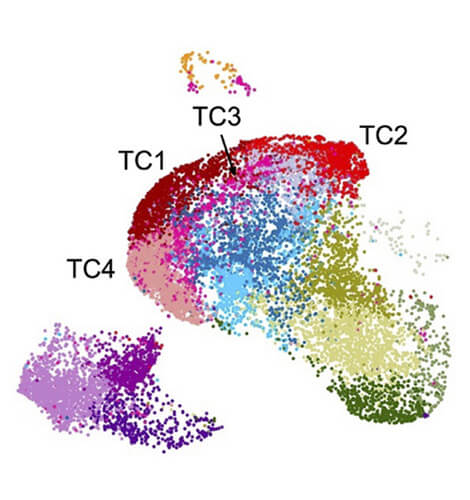

RNA Biology and Posttranscriptional Regulation
Head: Prof. Markus Landthaler
The Landthaler lab is interested in system-wide understanding of posttranscriptional regulation in mammalian cells in response to changes in their immediate environment.
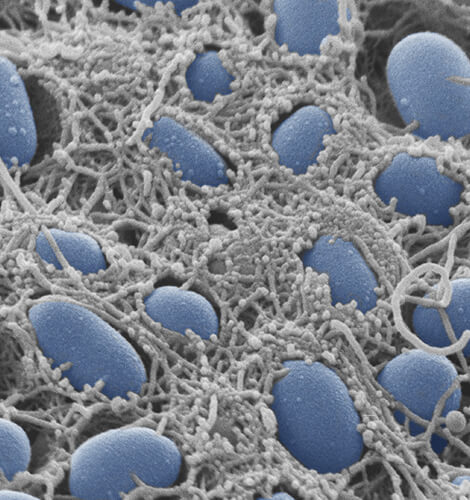

General Microbiology
Head: Prof. Regine Hengge
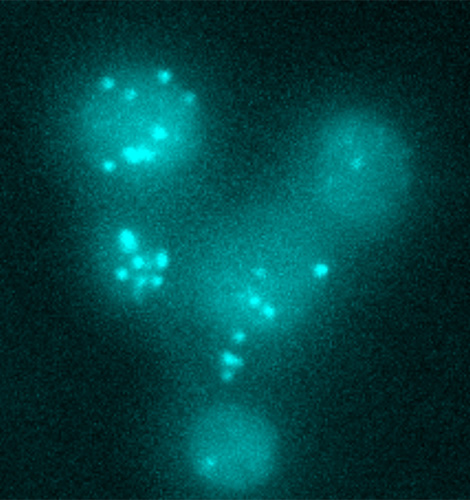

Theoretical Biophysics
Head: Prof. Edda Klipp
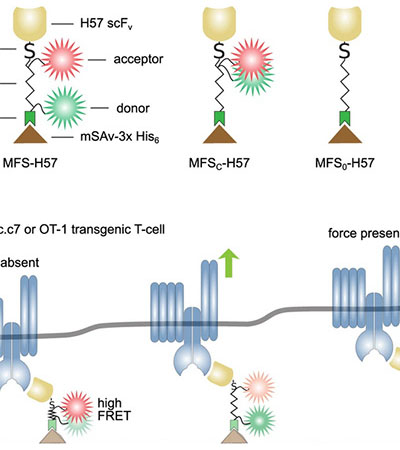

Experimental Biophysics / Mechanobiology
Head: Prof. Enrico Klotzsch
Our group aims in bridging the gap between structural and cell biology, particularly to decipher the mechanical aspects of how cells sense and react to their environment.

Experimental Biophysics
Head: Prof. Peter Hegemann
Our research group studies the structure-function relation of sensory photoreceptors from algae, fungi and bacteria.
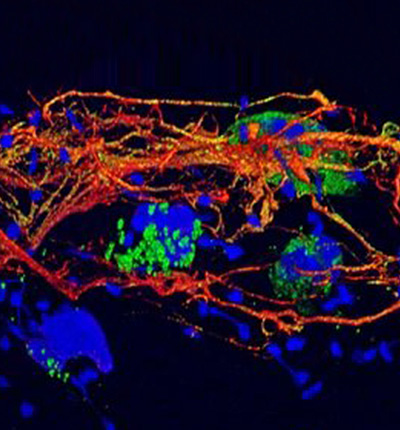
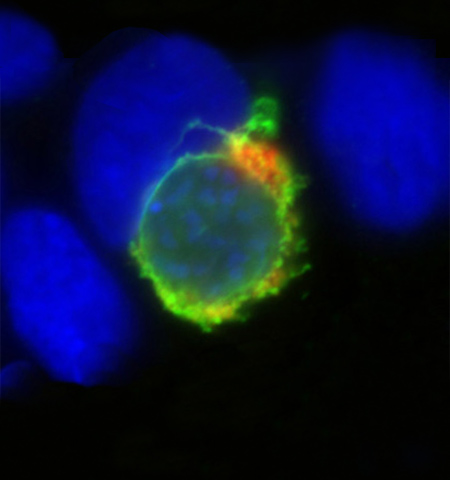

Molecular parasitology
Head: Prof. Kai Matuschewski

Molecular Genetics
Head: Prof. Christian Schmitz-Linneweber

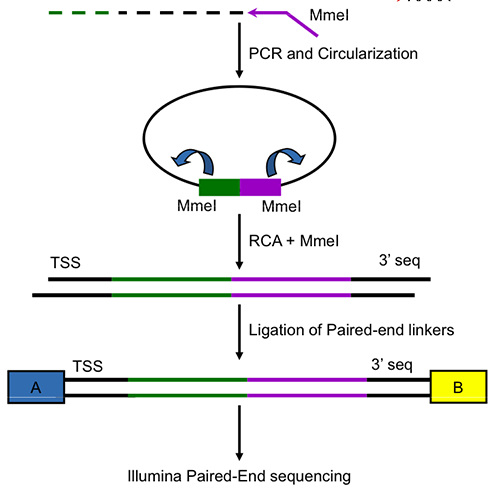

Computational regulatory genomics
Head: Prof. Uwe Ohler

Complex Systems
Head: Prof. Dirk Brockmann
Our research is focused on the development of mathematical and computational models for complex dynamical phenomena in the life-sciences.



Comparative Zoology
Head: Prof. John Nyakatura
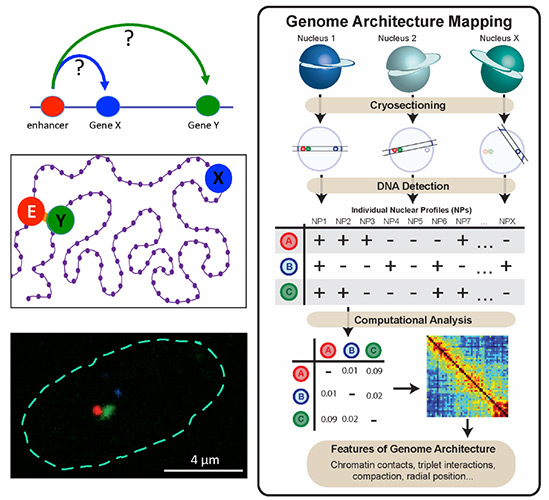

Transcriptional Regulation and Genome Architecture
Head: Prof. Ana Pombo

Microbiology
Head: Prof. Thomas Eitinger


Plant Evolution and Biodiversity
Head: Prof. Susann Wicke
We study interactions between plant parasites, their hosts, and the abiotic environment using experimental-genetic and evolutionary-ecological methods to understand the feedback loops driving parasite-host-environment adaptation. The lab develops resources to identify pests from environmental samples and engages in forest genetics research. We also actively commit to the conservation of native plants through citizen science projects.


Collective Information Processing
Head: Prof. Pawel Romanczuk
We investigate the interplay of self-organization and function in complex biological systems by combining mathematical modeling and analysis of experimental data. Our main research focus are mechanisms and functional benefits of collective behavior from cellular ensembles to animal groups. Further topics include modeling of stochastic motility and spatially-explicit evolutionary games.
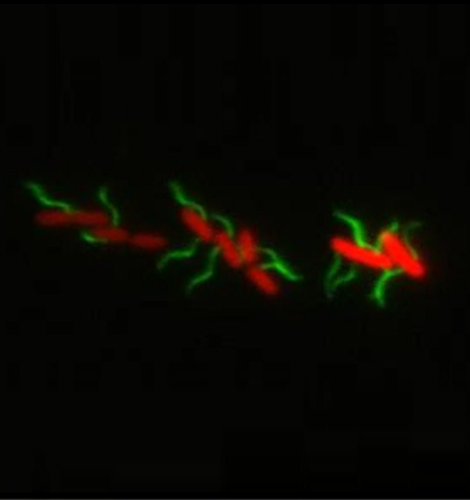

Molecular Microbiology
Head: Prof. Marc Erhardt
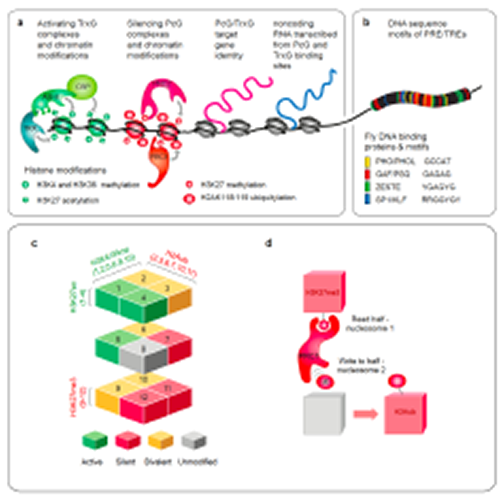
Quantitative Biology of the Eukaryotic Cell
Head: Prof. Leonie Ringrose






