Radical Enzymes
Radical enzymes use the high reactivity of protein and/or substrate-based radicals to convert inert compounds in the biosphere under anoxic (dioxygen depleted) conditions.
Fe/S cluster containing glycyl-radical enzymes (Fe/S-GREs) catalyze key steps in the activation of C-H bonds of aromatic and aliphatic compounds. Functional enzymes are generated by post-translational activation by a specific Fe/S activating enzyme belonging to the S-adenosylmethionine (SAM) radical superfamily.
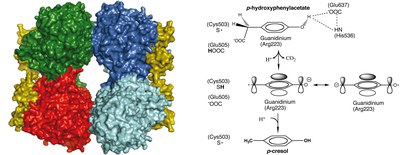
4-hydroxyphenylacetate decarboxlase (4Hpad) is a 2x[4Fe-4S] clusters GRE using a protein-based glycyl/thiyl radical dyad to catalyze the decarboxylation of 4-hydroxyphenylacetato into p-cresol (4-methylphenol), a virulence factor of the human pathogen Clostridium difficile.
The structural analyses of 4Hpad in both the substrate-free and substrate-bound states (BM Martins et al, JACS 2011), together with biochemical studies (Yu et al. Biochemistry 2006; Selmer et al. Eur J Biochem 2001) and theoretical calculations (M Feliks et al. JACS 2013) supports a Kolbe-type decarboxylation for p-cresol formation, an unprecedented reaction in biology.
The project was financed by the DFG SPP1319 "Biological Transformations of hydrocarbons without oxygen: from the molecular to the global sale".
Atypical dehydratases use radical intermediates to lower the pK of the non-activated beta-carbon hydrogen by 26 units thus facilitating the elimination of a water molecule (hydroxyl group eliminated from gamma-carbon).
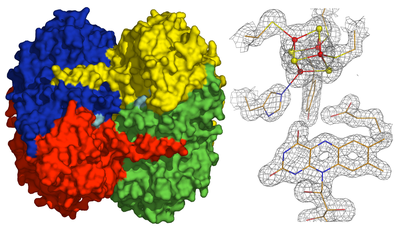
4-hydroxybutyryl-CoA dehydratase (4HDB) catalyzes the reversible dehydration of 4-hydroxybutyryl-CoA by a ketyl radical mechanism yielding crotonyl-CoA, the key step in the fermentation of 4-aminobutyrate in Clostridium aminobutyricum. 4HBD was identified as one of the central enzymes involved in global carbon cycling (Berg et al, Science 2007).
The crystal structure shows an unprecedented active site with a [4Fe-4S] cluster, coordinated by three cysteine and one histidine residues, located 7 Å from the FAD moiety. The substrate can bind between the [4Fe-4S] cluster and the FAD with both cofactors contributing to its activation and catalytic conversion.
The project was financed by the DFG SPP1071 "Radicals in enzymatic catalysis".
