Publications
Richter, F., Scheib, U., Mehlhorn, J., Schubert, R., Wietek, J., Gernetzki, O., Hegemann, P., Mathes, T., Möglich, A. (2015) Upgrading a Microplate Reader for Photobiology and All-Optical Experiments. Photochem. Photobiol. Sci. in press [PDF] [Scopus(0)]
Gasser, C. F., Taiber, S., Yeh, C.-M., Wittig, C. H., Hegemann, P., Ryu, S., Wunder, F., Möglich, A. (2014) Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc. Natl. Acad. Sci. U. S. A. 111(24), 8803-8808 [PDF] [Scopus(8)]
Diensthuber, R. P., Engelhard, C., Lemke, N., Gleichmann, T., Ohlendorf, R., Bittl, R., Möglich, A. (2014) Biophysical, mutational and functional investigation of the chromophore-binding pocket of light-oxygen-voltage photoreceptors. ACS Synth. Biol. 3(11), 811-819 [PDF] [Scopus(0)]
Gleichmann, T., Diensthuber, R. P., Möglich, A. (2013) Charting the Signal Trajectory in a Light-Oxygen-Voltage Photoreceptor by Random Mutagenesis and Covariance Analysis. J. Biol. Chem. 288(41), 29345-29355 [PDF] [Scopus(3)]
Möglich, A., Hegemann, P. (2013) Biotechnology: Programming genomes with light. Nature 500(7463), 406-408 [PDF] [Scopus(1)]
Engelhard, C., Raffelberg, S., Tang, Y., Diensthuber, R. P., Möglich, A., Losi, A., Gärtner, W., Bittl, R. (2013) A structural model for the full-length blue light-sensing protein YtvA from Bacillus subtilis, based on EPR spectroscopy. Photochem. Photobiol. Sci. 12(10), 1855-1863 [PDF] [Scopus(2)]
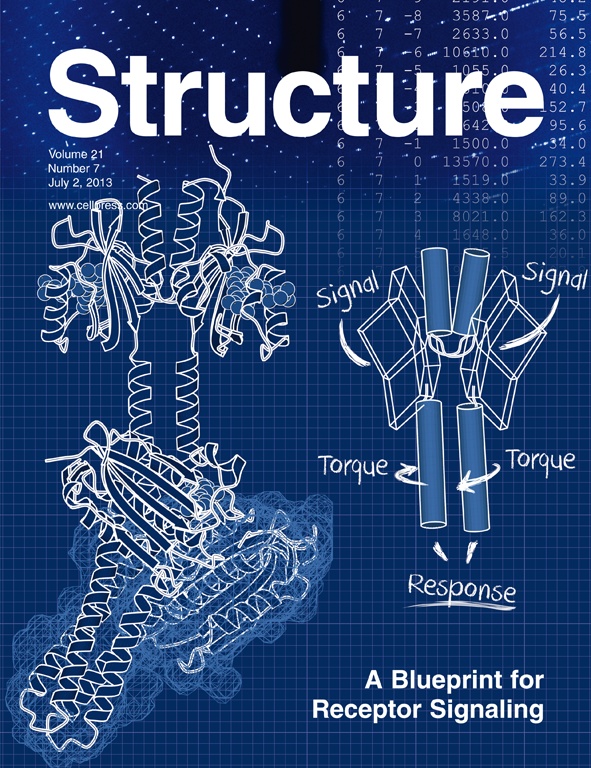 Diensthuber, R. P., Bommer, M., Gleichmann, T., Möglich, A. (2013) Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21(7), 1127-1136 [PDF] [Scopus(24)]
Diensthuber, R. P., Bommer, M., Gleichmann, T., Möglich, A. (2013) Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21(7), 1127-1136 [PDF] [Scopus(24)]
Diensthuber, R. P., Ohlendorf, R., Gleichmann, T., Schubert, R., Möglich, A. (2013) Lichtregulierte Genexpression. BioSpektrum 19(2), 149-151 [PDF] [Scopus(0)]
Rockwell, N. C., Ohlendorf, R., Möglich, A. (2013) Cyanobacteriochromes in full color and three dimensions. Proc. Natl. Acad. Sci. U. S. A. 110(3), 806-807 [PDF] [Scopus(4)]
Ohlendorf, R., Vidavski, R. R., Eldar, A., Moffat, K., Möglich, A. (2012) From Dusk Till Dawn: One-Plasmid Systems for Light-regulated Gene Expression. J. Mol. Biol. 416(4), 534-542 [PDF] [Scopus(27)]
Hegemann, P., Möglich, A. (2011) Channelrhodopsin engineering and exploration of new optogenetic tools. Nature Methods 8(1), 39-42 [PDF] [Scopus(49)]
Möglich, A., Moffat, K. (2010) Engineered Photoreceptors as Novel Optogenetic Tools. Photochem. Photobiol. Sci. 9(10), 1286-1300 [PDF] [Scopus(79)]
Möglich, A., Ayers, R. A., Moffat, K. (2010) Addition at the Molecular Level: Signal Integration in Designed Per-ARNT-Sim Receptor Proteins. J. Mol. Biol. 400(3), 477-486 [PDF] [Scopus(40)]
Möglich, A., Yang, X., Ayers, R. A., Moffat, K. (2010) Structure and Function of Plant Photoreceptors. Annu. Rev. Plant Biol. 61, 21-47 [Link] [Scopus(114)]
Möglich, A., Ayers, R. A., Moffat, K. (2009) Structure and Signaling Mechanism of Per-Arnt-Sim Domains. Structure 17(10), 1282-1294 [PDF] [Scopus(150)]
Möglich, A., Ayers, R. A., Moffat, K. (2009) Design and Signaling Mechanism of Light-Regulated Histidine Kinases. J. Mol. Biol. 385(5), 1433-1444 [PDF] [Scopus(119)]
Möglich, A., Moffat, K. (2007) Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol. 373(1), 112-126 [PDF] [Scopus(102)]
Möglich, A., Joder, K., Kiefhaber, T. (2006) End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Natl. Acad. Sci. U. S. A. 103(33), 12394-12399 [Scopus(142)]
Möglich, A., Krieger, F., Kiefhaber, T. (2005) Molecular basis for the effect of urea and guanidinium chloride on the dynamics of unfolded polypeptide chains. J. Mol. Biol. 345(1), 153-162 [Scopus(79)]
Krieger, F., Möglich, A., Kiefhaber, T. (2005) Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J. Am. Chem. Soc. 127(10), 3346-3352 [Scopus(78)]
Möglich, A., Weinfurtner, D., Gronwald, W., Maurer, T., Kalbitzer, H. R. (2005) PERMOL: restraint-based protein homology modeling using DYANA or CNS. Bioinformatics 21(9), 2110-2111 [Scopus(5)]
Möglich, A., Weinfurtner, D., Maurer, T., Gronwald, W., Kalbitzer, H. R. (2005) A restraint molecular dynamics and simulated annealing approach for protein homology modeling utilizing mean angles. BMC Bioinformatics 6, 91 [Scopus(9)]
Güthe, S., Kapinos, L., Möglich, A., Meier, S., Grzesiek, S., Kiefhaber, T. (2004) Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J. Mol. Biol. 337(4), 905-915 [Scopus(34)]
Martin, G., Möglich, A., Keller, W., Doublié, S. (2004) Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 341(4), 911-925 [Scopus(36)]
Möglich, A., Koch, B., Gronwald, W., Hengstenberg, W., Brunner, E., Kalbitzer, H. R. (2004) Solution structure of the active-centre mutant I14A of the histidine-containing phosphocarrier protein from Staphylococcus carnosus. Eur. J. Biochem. 271(23-24), 4815-4824 [Scopus(8)]
Gröger, C., Möglich, A., Pons, M., Koch, B., Hengstenberg, W., Kalbitzer, H. R., Brunner, E. (2003) NMR-spectroscopic mapping of an engineered cavity in the I14A mutant of HPr from Staphylococcus carnosus using xenon. J. Am. Chem. Soc. 125(29), 8726-8727 [Scopus(27)]
Möglich, A., Wenzler, M., Kramer, F., Glaser, S. J., Brunner, E. (2002) Determination of residual dipolar couplings in homonuclear MOCCA-SIAM experiments. J. Biomol. NMR 23(3), 211-219 [Scopus(6)]
Book chapter
Moffat, K., Zhang, F., Hahn, K., Möglich, A. (2013) The Biophysics and Engineering of Signaling Photoreceptors. In Optogenetics (edited by Hegemann & Sigrist), De Gruyter, Berlin, Germany.
