Research
Research in our laboratory mainly focuses on the transcription of plastid and mitochondrial genomes and on post-transcriptional processes in these genetically semi-autonomous, energy-converting organelles. We are additionally studying mitochondrial biogenesis and function in plants. Most of our work is done using the model plant Arabidopsis thaliana.
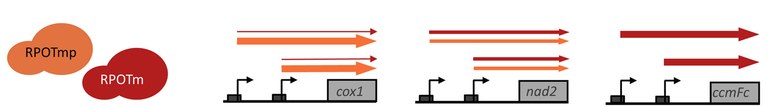
(Model for gene-specific transcriptional tasks of the phage-type RNA polymerases RPOTm and RPOTmp in mitochondria)
Our research uses
- Transgenic plants for reverse-genetic approaches
- Various techniques for high- and low-throughput analyses of organelle transcripts and for transcript end mapping
- Cell fractionation techniques
- In organello assays for monitoring mitochondrial transcription and translation
- Native PAGE and respiration assays for studying the mitochondrial oxidative phosphorylation system
Currently funded projects:
Mechanisms controlling transcription of the plastid genome (FP7 Marie Curie Career Integration Grant POLSPEC)
Organellar phage-type RNA polymerases are indispensable for the transcription of the chloroplast genome. They have fundamental roles in the biogenesis of the photosynthetic compartment of plant cells. The project POLSPEC investigates distinct roles of two phage-type RNA polymerases, RPOTp and RPOTmp, which are present in plastids of dicotyledonous plants where they transcribe the plastid genome together with a third, eubacterial-type transcriptase named PEP. The project has two major objectives: 1. Defining the RPOTp- and RPOTmp-specific plastid transcriptomes and generating a plastid genome-wide map of transcription start sites (TSS) utilised by RPOTp or RPOTmp. 2. Identifying cis-regulatory elements on the plastid genome that direct the transcriptional activities of RPOTp and RPOTmp.


Chloroplast RNA polymerase activity during acclimation (DFG TRR 175 project A01)
This project aims to quantify transcriptional alterations that occur in chloroplasts during acclimation to changes in light or temperature. It will apply chloroplast nascent transcript sequencing strategies (GRO-seq and 5’-GRO-seq) in order to globally assess transcriptional contributions to changes in chloroplast gene expression during acclimation. By combining high-resolution quantitative transcriptional analyses with a reverse genetic approach we will determine distinct contributions of individual chloroplast RNA polymerases to transcriptional alterations.
See TRR 175 for more information.
![]()

