Mass Spectrometry
“Atomic particles stored in a cage without material walls”
GC-MS with a Single Quadrupole
GC-FID is the most commonplace analyses in lipid research, however additional confirmation of analytes is essential in complex biological samples. We use qualitative analysis of fatty acid methyl esters (FAMEs) by gas chromatography-mass spectrometry (GC-MS) for correct identification of chromatographic peaks and determination of co-elution.
We run both EI and CI ion sources. Electron impact ionisation (EI) is the standard process in all GC-MS instruments. In EI molecular ions M+ (generated by loss of an electron) are produced by bombardment with high energy electrons, resulting in fragmentation and specific M+ pattern. In contrast chemical ionisation (CI) is a soft ionisation technique, which involves an exothermic chemical reaction by a reagent gas and its reagent ions. CI spectra therefore have few or no fragments and thus give information on the molecule itself. We use CI for structure determination and confirmation of molecular weights in hydroxy-FAMEs.

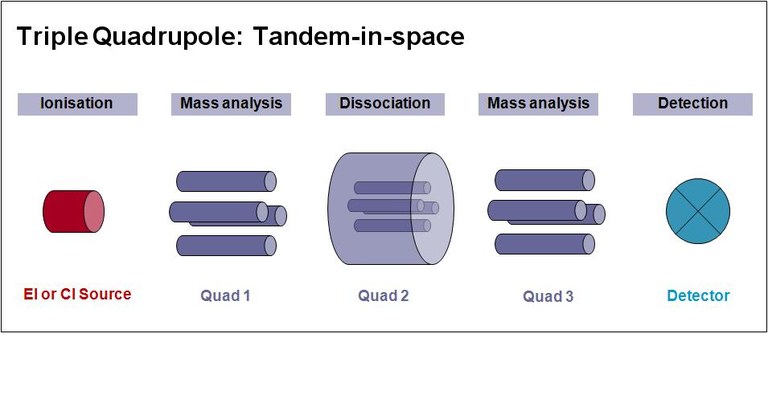
GC-MS/MS using a Triple Quadrupole (QQQ)
In MS/MS or tandem MS analytical steps take place in different locations in the beam path of the instrument. The basic QQQ design consist of three quadrupoles in a row, of which Q1 and Q3 act as mass filter, whereas the middle one (Q2) is used as collision cell. In Q1 the precursor ion is selected and afterwards dissociated into product ions in Q2. This process is followed by a mass scan in Q3.
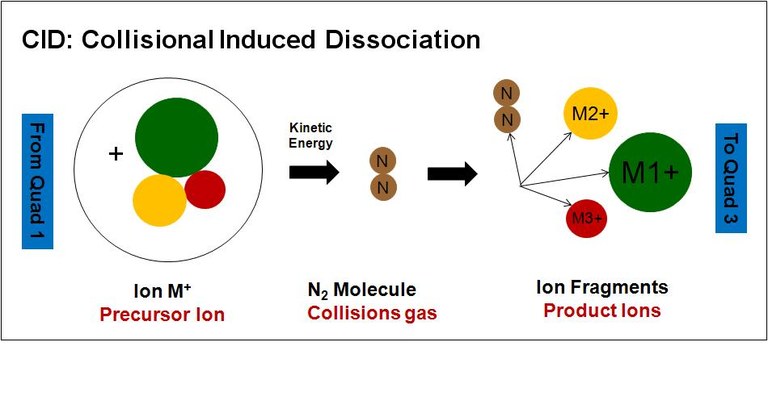
In the collision cell Collision Induced Dissociation (CID) of the precursor ion is performed. Energy is transformed through collision with neutral molecules (collision gas), resulting in bond cleavages and rearrangements of the selected ion. Key fragmentation mechanisms are used to interpret CID mass spectra.
Why do we use MS/MS?
Firstly, we work with lipid extract derived from complex media such as soil. Due to this the sample matrix contributes significantly to the chemical background noise. Moreover, analysis is often hampered by co-elution of isobaric impurities. With MS/MS we separate target from matrix and significantly improve the signal to noise ratio. Secondly, soil organisms are very low in biomass and MS/MS allows us detection of FAMEs in the femto gram range. In sum, for us the main benefit of MS/MS over classical MS is identification and quantitation with the highest possible sensitivity in difficult matrices.