.
C-di-GMP signaling in E. coli biofilm formation
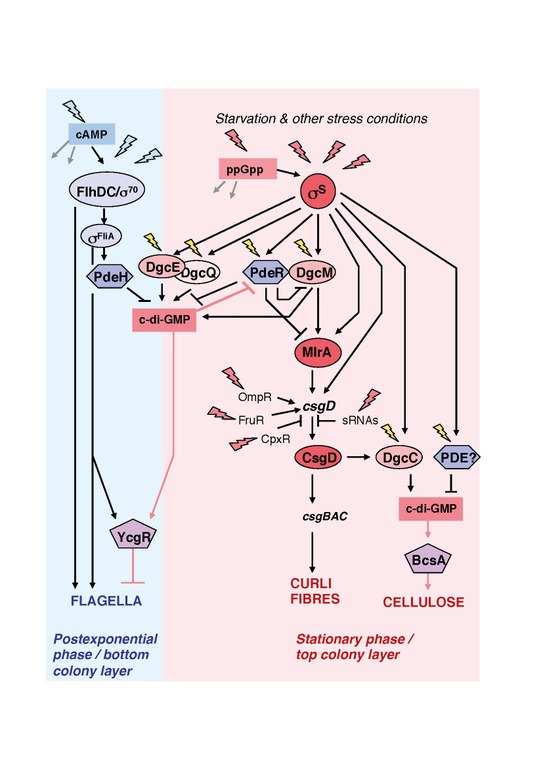
C-di-GMP is a ubiquitous bacterial nucleotide second messenger, which in general down-regulates flagellar expression or activity and promotes biofilm-related functions such as the production of matrix components or adhesins. In pathogenic bacteria it often also controls virulence genes. In E. coli the production of amyloid curli fibres and cellulose is promoted by c-di-GMP, which is synthesized by 12 diguanylate cyclases (DGC, with GGDEF domains) and degraded by 13 phosphodiesterases (PDE, with EAL domains). Some of these DGCs and PDEs have very specific output functions, suggesting some kind of 'local' c-di-GMP signaling.
Several of these DGCs (DgcE, DgcM) and PDEs (PdeH, PdeR) and the MerR-like transcription factor (TF) MrlA specifically regulate the transcription of csgD, which encodes a biofilm regulator essential for producing amyloid curli fibres and cellulose as a biofilm matrix. This system operates as a signaling cascade, in which c-di-GMP controlled by the DGC/PDE pair DgcE/PdeH (module I) regulates the activity of the DgcM/PdeR pair (module II). Via multiple direct interactions, the two module II proteins interact with MlrA. PdeR acts as a connector between modules I and II and operates as a trigger enzyme: its direct inhibition of the DGC DgcM and the TF MlrA is relieved when it binds and degrades c-di-GMP generated by module I. As a consequence, DgcM then also generates c-di-GMP which provides for a positive feedback. In addition, DgcM – by direct and specific interaction – activates MlrA and RpoS-RNAP to initiate csgD transcription. CsgD then turns on the curli genes as well as dgcC, which encodes a DGC that specifically activates cellulose synthase. Overall, the combination of DGC/PDE activity with highly specific regulatory protein-protein interactions, is likely to represent a general principle in local c-di-GMP signaling.
In addition, most of the other DGCs and PDEs are expressed but do not contribute to this scenario under standard laboratory conditions, suggesting that these enzymes are present in an inactive form. However, specific signals perceived by the various N-terminal sensor domains may activate these DGCs and PDEs and thereby modulate c-di-GMP levels, matrix production and biofilm morphology.
Questions currently addressed in our group include the following:
- Are there additional PDEs like PdeR, that act as 'trigger enzymes' in local c-di-GMP signaling?
- How do the DGC and co-activator DgcM, the TF MlrA, additional TFs and RpoS-RNAP cooperate to activate the csgD promoter?
- How does c-di-GMP control via PdeR/DgcM/MlrA contribute to bistable CsgD expression in zones of heterogeneous matrix production in macrocolony biofilms?
- How does the DGC DgcC specifically activate cellulose synthase?
- What are the signals sensed by the diverse N-terminal sensory domains of the E. coli DGCs and PDEs? What are the molecular mechanisms of this sensory input?
- Are there still unknown c-di-GMP-binding effector proteins in E. coli?
Herbst, S., M. Lorkowski, O. Sarenko, T.K.L. Nguyen, T. Jaenicke, and R. Hengge (2018) Transmembrane redox control and proteolysis of PdeC, a novel type of c-di-GMP phosphodiesterase. EMBO J. 37: e97825 (published ahead of print on Apr 13).
Thongsomboon, W., D.O. Serra, A. Possling, C. Hadjineophytou, R. Hengge and L. Cegelski (2018) Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science 359: 334-338. doi: 10.1126/science.aao4096.
Sarenko, O., G. Klauck, F.M. Wilke, V. Pfiffer, A.M. Richter, S. Herbst, V. Kaever, and R. Hengge (2017) More than enzymes that make and break c-di-GMP - the protein interaction network of GGDEF/EAL domain proteins of Escherichia coli. mBio 8, e01639-17.
Hengge, R. (2016) Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins. Phil. Trans. R. Soc. B 371: 20150498, doi:
Hengge, R., M.Y. Galperin, J.-M. Ghigo, M. Gomelsky, J. Green, K.T. Hughes, U. Jenal, and P. Landini (2015) Systematic nomenclature for GGDEF and EAL domain-containing c-di-GMP turnover proteins in Escherichia coli. J. Bacteriol., 98, 7-11. doi: 10.1128/JB.00424-15.
Povolotsky, T.L., and R. Hengge (2015) Genome-based comparison of c-di-GMP signaling in pathogenic and commensal Escherichia coli strains. J. Bacteriol., 198,111-26, doi: 10.1128/JB.00520-15.
Lindenberg, S., G. Klauck, C. Pesavento, H. Weber, E. Klauck, and R. Hengge (2013) The EAL protein YciR is a trigger enzyme in a c-di-GMP signaling cascade in E. coli biofilm control. EMBO J. 32, 2001-2014.
Hengge, R. (2009) Principles of cyclic-di-GMP signaling. Nature Rev. Microbiol. 7, 263-273.
Tschowri, N., S. Busse, and R. Hengge (2009) The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue light stress response of E.coli. Genes Dev. 23, 522-534.
Sommerfeldt, N., A. Possling, N. Tschowri, G. Becker, C. Pesavento, H. Weber, and R. Hengge (2009) Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155, 1318-1331.
Pesavento, C., G. Becker, N. Sommerfeldt, A. Possling, N. Tschowri, A. Mehlis, and R. Hengge (2008) Inverse regulatory coordination of motility and curli-mediated adhesion of E.coli. Genes Dev. 22, 2434-2446.
Weber, H., C. Pesavento, A. Possling, G. Tischendorf, and R. Hengge (2006) Cyclic-diGMP signaling within the sS network of Escherichia coli. Mol. Microbiol. 62, 1014-1034.