Blue-Light Receptors
|
Blue-light sensitive photoreceptors using flavin chromophores (Figure 1) control many crucial biological processes. In contrast to classical photoreceptors using isomerizing cofactors like carotenoids or tetrapyrrols these photoreceptors use typical flavin(bio-)chemistry to facilitate light-induced signaling. |
||
|
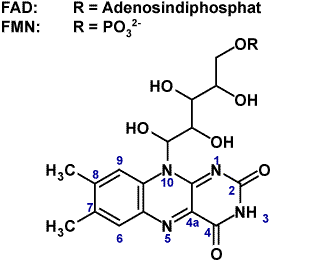
Blue-light biological receptor groups containing flavin-adenin-dinucleotide (FAD) are photolyases, cryptochromes (Cry) and photoreceptors with BLUF (blue light sensors using FAD)-domains.The blue-light sensitive phototropins (phot) use LOV (Light-Oxygen-Voltage) domains that bindflavin-mononucleotide (FMN). The phototropins and homologues (phot-proteins) are involved in phototropism (plant growth towards light source), chloroplast movement, stomata opening (in guard cells), rapid inhibition of stem growth, and gametogenesis (sexual differentiation in algae). Receptors with BLUF-domains are functioning as redox and light-regulated de-repressors in purple bacteria or as photoreceptors for behavioral responses in flagellates like Euglena. Additionally they regulate biofilm formation and virulence in (pathogenic) non-photosynthetic organisms. |
Figure1, Riboflavin is the common component of blue light receptors.
|
|
|
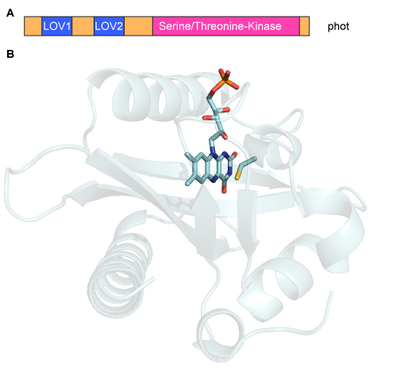
1. Similar to plants the unicellular algae Chlamydomonasreinhardtiiharbors aphot-receptor (CrPhot). Unlike in plants it is involved in sexual reproduction and regulation of photosynthesis genes. We use CrPhot as a phot model system to study structure/function relationships of photsignaling processes. Wild type proteins or protein domains are expressed in Escherichia coli. |
||
|
Figure2, Domain arrangement of photproteins (A) and structure of a LOV domain (B). |
The purified proteins and especially the chromophore-binding domains are studied by absorption and fluorescent spectroscopy, flash-photolysis, time resolved fluorescence (A. Penzkofer, Regensburg university) [1], infrared spectroscopy (T. Kottke, Bielefeld University)[2], and electron spin resonance spectroscopy (EPR, R.Bittl (FU Berlin) and S.Weber/E. Schleicher (Freiburg University) [3]. Atomic structures are solved in collaboration with I. Schlichting (Max Planck institute for biomedical research, Heidelberg) [4]. Additionally we study phototropin function in vivo using phot knock-out strains of Chlamydomonas [5]. The combination of high-resolution structural models with time-resolved spectroscopic data and in vivo analysis will finally elucidate the molecular signal transductionmechanism. |
|
|
2. Photoactivated adenylyl cyclases (PAC) employ BLUF domains to regulate the activity of adenylyl cyclase domains and are therefore highly interesting optogenetic tools to manipulate cAMP induced responses in vivo. |
||
|
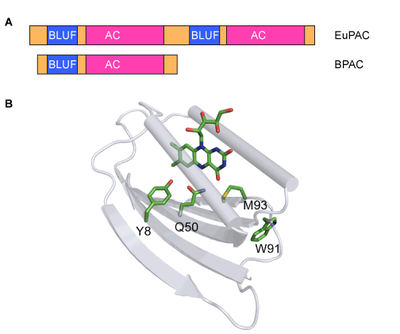
The first described PAC was found in Eulenagracilis (EuPAC) and is composed of a complex tetramer formed by two photo-activated adenylatecyclases (PACa and PACb).Both subunits contain two BLUF domains and two cyclase domains (AC) (Figure 3). Recently, a bacterial homolog (bPAC) was found in Beggiatoa sp., which is of much simpler design.[6] The homodimer consists of one BLUF and one AC domain per monomer and is superior to EuPAC in terms of size, activity and dynamic range. With the ultimate goal to rationally design novel PACs with customized activity and sensitivity we study the interaction of BLUF sensor and AC effector domains using biochemical and electrophysiological assays, crystallography and spectroscopy. |
Figure3, Domain architecture of PACs (A) and structure of a BLUF domain (B). |
|
In this regard we also study the internal signal propagation in BLUF domains using high-end time resolved vibrational spectroscopy and selective isotope labeling.
References:
1. Holzer W., Penzkofer A., Susdorf T., Alvarez M., Islam S. D. M., Hegemann P. 2004 Absorption and emission spectroscopic characterisation of the LOV2-domain of phot from Chlamydomonas reinhardtii fused to a maltose binding protein. Chemical Physics, 302(1-3), 105-118.
2. Pfeifer A., Mathes T., Lu Y., Hegemann P., Kottke T. 2010 Blue light induces global and localized conformational changes in the kinase domain of full-length phototropin. Biochemistry, 49(5), 1024-1032.
3. Schleicher E., Kowalczyk R. M., Kay C. W. M., Hegemann P., Bacher A., Fischer M., Bittl R., Richter G., Weber S. 2004 On the reaction mechanism of adduct formation in LOV domains of the plant blue-light receptor phototropin. Journal of the American Chemical Society, 126(35), 11067-11076.
4. Fedorov R., Schlichting I., Hartmann E., Domratcheva T., Fuhrmann M., Hegemann P. 2003 Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii. Biophysical Journal, 84(4), 2474-2482.
5. Trippens J., Greiner A., Schellwat J., Neukam M., Rottmann T., Lu Y., Kateriya S., Hegemann P., Kreimer G. 2012 Phototropin Influence on Eyespot Development and Regulation of Phototactic Behavior in Chlamydomonas reinhardtii. Plant Cell.
6. Stierl M., Stumpf P., Udwari D., Gueta R., Hagedorn R., Losi A., Gärtner W., Petereit L., Efetova M., Schwarzel M., et al. 2011 Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem, 286(2), 1181-1188.
7. Bonetti C., Mathes T., van Stokkum I. H. M., Mullen K. M., Groot M. L., van Grondelle R., Hegemann P., Kennis J. T. M. 2008 Hydrogen Bond Switching among Flavin and Amino Acid Side Chains in the BLUF Photoreceptor Observed by Ultrafast Infrared Spectroscopy. Biophysical Journal, 95(10), 4790-4802.
8. Mehlhorn J., Steinocher H., Beck S., Kennis J. T., Hegemann P., Mathes T. 2013 A Set of Engineered Escherichia coli Expression Strains for Selective Isotope and Reactivity Labeling of Amino Acid Side Chains and Flavin Cofactors. PLOS one, 8(11), e79006.