Genome Editing in Chlamydomonas reinhardtii
Support our iGEM team: https://www.startnext.com/igemhu2019/ Visit their official wiki here: https://2019.igem.org/Team:Humboldt_Berlin
You can find a list of our mutants strains here (chlamy.de/strains)
Our lab supports the first HU iGEM team. The students set out to solve the microplastic problem by creating green algae capable of degrading plastic. Follow their updates. For more info visit www.igem.hu-berlin.de, to become a sponsor or to get involved contact igem@chlamy.de.
Our Chlamy team will attend the 18th Chlamy conference in Washington, D.C. Looking forward to many exciting discussions and inspiring talks and posters!
CC-125 and SAG73.72 cells were electroporated using one 8-ms/ 300-V poring pulse, followed by five 50-ms/20-V polarity-exchanged transfer pulses at 50-ms intervals and a decay rate of 40%.
You can find a list of all our CRISPR/Cas9 plasmids here (chlamy.de/plasmids) and the first photoreceptor mutants here (chlamy.de/strains)
Here is nice feature from the rbb about the chlamylicious iGEM project:
(c) rbb
The Chlamy group in the Hegemann lab is interested in the characterization of known but so far uncharacterized photoreceptors and newly discovered ones. Many of these photoreceptor proteins were found in the model alga Chlamydomonas reinhardtii. In addition to in vitro electrophysiological and spectroscopic characterizations, we aim to decipher the photoreceptor network in Chlamydomonas.
Our incentive is to delete the photoreceptors separately and in total and to specifically modify them to understand their biochemical function and physiological role.
Team
Heide Evers, Oga Baidukova, Simon Kelterborn, Yousef Kamrani, Dr. Irina Sizova, Dimitri Schuhmacher and Darius Rauch.
Contact us:
genome-engineering@chlamy.de
Resources
- Join the discussion on ResearchGate.
- http://www.chlamycollection.org/ Great resources for Chlamydomonas strains and plasmids.
- Our CRISPR/Cas9 plasmids and CRISPR/Cas9 mutants are now available at the Chlamydomonas Resource Center.
- For recombinant Cas9 protein expression in E. coli, we use plasmid pET-28b-Cas9-His (addgene #47327) of the Alex Schier Lab and their protocol for purification.
- Our favourite CRISPR online tool for gRNA design and OFF-target prediction supporting the Chlamydomonas reinhardtii genome: CRISPR-P.
- How to find light in the dark: https://phototaxis.bandcamp.com/
Project Background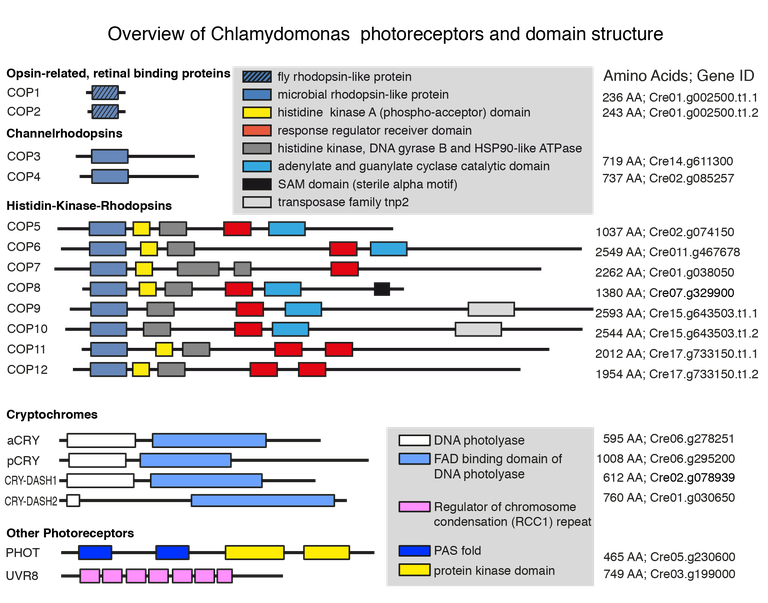
Figure 1: Overview of predicted photoreceptors (Greiner et al. 2017)
DNA double-strand breaks (DSBs) in the gene of interest are prerequisites for capitalizing the cells own DNA repair mechanism that ultimately results in gene deletion or modification. Widely used programmable site-directed DNA-cleaving enzymes are zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN) and the CRISPR/Cas9 system (Clustered Regularly Interspaced Short Palindromic Repeats). Cells have different mechanisms to cope with DSB. Repair of DNA lesions by non-homologous end-joining (NHEJ) is error-prone and lead to unpredictable modifications on the target site. In contrast, homologous recombination (HR) allows the defined deletion (knock out), repair (rescuing), tagging (knock-in) and modification of selected genes. However, HR is an extremely inefficient process in most eukaryotes including algae, plants, and animals
Targeted genome editing with programmable nucleases
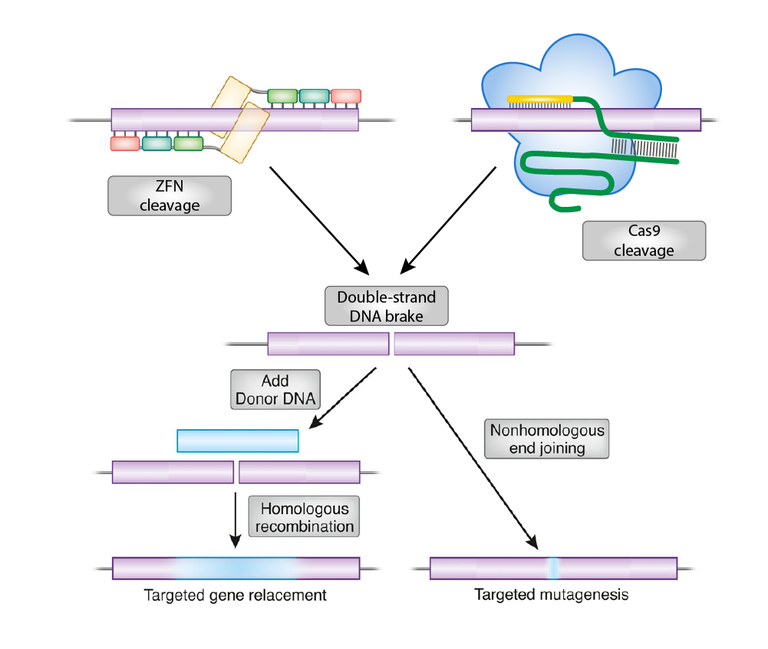
Figure 2: Genome editing with programmable nucleases. Adapted from Dana Carroll (2011)
In Chlamydomonas reinhardtii, HR efficiency was greatly enhanced by introducing double-strand breaks with the help of two engineered zinc-finger nuclease (ZFN) proteins (Sizova et al. 2013).
Now, we further optimized the protocol to target genes also in motile strains and adapted to use the CRISPR/Cas9 system (Greiner et al. 2017).
Furthermore, we are looking for genes influencing the transformation, gene targeting efficiency, and DNA repair pathways, either by deletion or suppression of relevant proteins. The understanding of the underlying DNA repair mechanisms is fundamental to exploit this system and use it for precise genome editing experiments.
Knock-outs available from our lab
| Gene Name | Phytozome ID | Nuclease used for disruption | |
| COP1/2 | Cre01.g002500 | ZFN & Cas9 |
|
| ChR1 (COP3) | Cre14.g611300 | ZFN & Cas9 | |
| ChR2 (COP4) | Cre02.g085257 | ZFN & Cas9 | |
| ChR1 (COP3) + ChR2 (COP4) | ZFN & Cas9 | ||
| COP5 (HKR1) | Cre02.g074150 | Cas9 |
|
| COP6 (HKR2) | Cas9 |
|
|
| COP8 (HKR4) | Cas9 |
|
|
| COP11/12 (HKR7/8) | Cas9 |
|
|
| aCRY | Cre06.g278251 | Cas9 |
|
| Phot | Cre03.g199000 | Cas9 |
|
| UVR8 | Cre05.g230600 | Cas9 | |
| VGCC | Cre02.g074150 | Cas9 |
|
Contact for more information: genome-engineering@chlamy.de
Publications
Greiner A*, Kelterborn S*, Evers H, Kreimer G, Sizova I, and Hegemann P. (2017). Targeting of photoreceptor genes via zinc-finger nucleases and CRISPR/Cas9 in Chlamydomonas reinhardtii. The Plant Cell 29(10). [DOI: 10.1015/tpc17.00659]
Hahn F, Mantegazza O, Greiner A, Hegemann P, Eisenhut M, & Weber A P M (2017). An Efficient Visual Screen for CRISPR/Cas9 Activity in Arabidopsis thaliana. Frontiers in Plant Science, 8(January), 39. [DOI: 10.3389/fpls.2017.00039]
Petroutsos D, Tokutsu R, Maruyama S, Flori S, Greiner A, Magneschi L, Minagawa J (2016). A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature, 537(7621), 563–566. [DOI: 10.1038/nature19358]
Sizova I, Greiner A, Awasthi M, Kateriya S, & Hegemann P (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. The Plant Journal : For Cell and Molecular Biology, 73(5), 873–82. [DOI: 10.1111/tpj.12066]
Trippens J, Greiner A, Schellwat J, Neukam M, Rottmann T, Lu Y, … Kreimer G (2012). Phototropin Influence on Eyespot Development and Regulation of Phototactic Behavior in Chlamydomonas reinhardtii. The Plant Cell, 24(11), 4687–4702. [DOI: 10.1105/tpc.112.103523]
Zorin B, Lu Y, Sizova I, Hegemann P. (2009). Nuclear gene targeting in Chlamydomonas as exemplified by disruption of the PHOT gene. Gene, 432(1–2), 91–6. [DOI: 10.1016/j.gene.2008.11.028]
Zorin B, Hegemann P, Sizova I (2005). Nuclear-gene targeting by using single-stranded DNA avoids illegitimate DNA integration in Chlamydomonas reinhardtii. Eukaryotic Cell, 4(7), 1264–72. [DOI: 10.1128/EC.4.7.1264-1272.2005]
Sizova I, Fuhrmann M, Hegemann P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene, 277(1–2), 221–229. [DOI: 10.1016/S0378-1119(01)00616-3]
 |
 |
Tags: www.chlamy.de CRISPR/Cas9 in Chlamydomonas Chlamy Chlamydomonas reinhardtii CRISPR/Cas9 Cas9 gRNA sgRNA Cpf1 gene editing genome engineering ZFN HR NHEJ