Crystallization
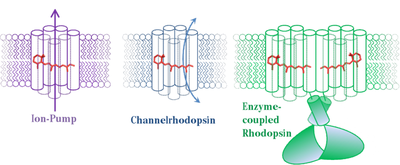
Rhodopsins are light responsive proteins, harboring a retinal cofactor as chromophore. They are found in all three domains of life. While the “classical” animal Rhodopsin in the photoreceptor cell of the eye represents prototypical G-protein coupled receptors, microbial Rhodopsins prevalently present in Achaea, Bacteria, and lower Eukaryotes show a wide variation of functionalities [1]. In the early 1970s , the first described member of this protein family is the light-driven proton-pump Bacteriorhodopsin from Halobacterium salinarum [2]. Since then many other, distinct microbial type Rhodopsins have been found, acting as ion pumps, light-gated ion channels, and light sensors [1]. Our group focuses on the investigation of structure-function relationship of light-gated ion-channels (Channelrhodopsins (ChR)) and the recently described class of enzyme-coupled Rhodopsins in particular. The vast progress made on the structural elucidation of ion pumping Rhodopsins over the past decades, allowed for a detailed understanding of the biophysical principles behind. However, the structural information regarding Channelrhodopsins or enzyme-coupled Rhodopsins (e.g. histindine-kinas-Rhodopsin (HKR) or Rhodopsin-cyclase (RhGC)) is rather rare or missing [3-5]. To overcome this limitation and gain insights into how the initial photochemical reaction triggers structural rearrangements within the protein we are working on solving the crystal structure of Channelrhodopsin-variants derived from algae as well as from representative members of enzyme-coupled Rhodopsins.
Crystallization of integral membrane proteins in general continues to be a challenge. These proteins are difficult to express, they are highly hydrophobic and therefore require detergent micelles to be stabilized in solution. Various expression systems are used to obtain a protein sample suitable for crystallization, including the pichia pastoris and insect cell/baculovirus system.

Flask-shaker with a suspension culture of insect-cells expressing recombinant microbial Rhodopsin.
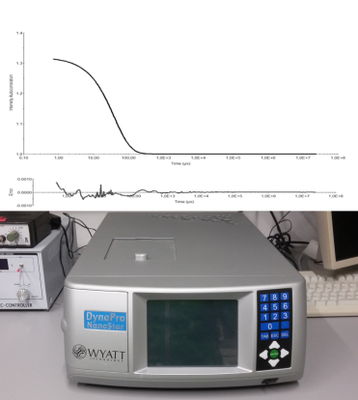
For protein purification the lab is equipped with two chromatography systems (ÄKTA pure and ÄKTA express, GE Healthcare). After purification dynamic light (DynaPro, Wyatt) scattering enable us to investigate the hydrodynamic behavior of the received sample to judge its suitability for crystallization.

Crystallization trials are set either following the “classical” vapour diffusion approach or in meso (lipidic cubic phase) using an Oryx 4 crystallization robot (Douglas Instruments). The latter method reconstitutes the protein in a bicontinuous lipid mesophase as crystallization matrix. This approach was very successfully implemented to obtain highly diffracting crystals from microbial Rhodopsin and GPCR [6].
Oryx 4 Crystallization Robot (Douglas Instruments Ltd.)
Several incubators allow for screening under distinct light and temperature conditions. Obtained crystals are harbored, cryoprotected in liquid nitrogen and tested for their x-ray diffraction at synchrotron sources (Bessy II, DESY or ESRF).

Protein-crystals derived from microbial Rhodopsins
The diffraction images collected from single crystals can be used to calculate the electron density map of the crystal unit cell which finally allows to model the three dimensional structure of the crystallized protein. Most crystallographic models are available from the protein data bank.
Involved in this project are:
Dr. Matthias Broser, Dr. Katja Stehfest, Melanie Meiworm
References:
[1] Ernst O., Lodowski D.T., Elstner M., Hegemann P., Brown L.S., Kandori H. (2014) "Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms" Chem. Rev., 114 (1): 126–163.
[2] Oesterhelt D.,Stoeckenius W. (1971) "Rhodopsin-like Protein from the Purple Membrane of Halobacterium halobium " Nature New Biology, 233: 149-152.
[3] Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, Ito J, Aita Y, Tsukazaki T, Hayashi S, Hegemann P, Maturana AD, Ishitani R, Deisseroth K, Nureki O (February 2012). "Crystal structure of the channelrhodopsin light-gated cation channel". Nature. 482 (7385): 369–74.
[4] Luck M, Mathes T., Bruun S., Fudim R., Hagedorn R., Nguyen T.,KateriyaS., Kennis J.T.M., Hildebrandt P.,Hegemann P. (2012) "A Photochromic Histidine Kinase Rhodopsin (HKR1) That Is Bimodally Switched by Ultraviolet and Blue Light" Journal of Biological Chemistry, 287(47): 40083–40090.
[5] Scheib U, Stehfest K, Gee CE, et al. The rhodopsin-guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Sci Signal. 2015;8(389):rs8.
[6] Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009).