RpoS dependent genes
Escherichia coli genes under the control of the general stress sigma factor σS (RpoS) identified by microarray analysis
A detailed description of the strains, procedures and microarrays used was published:
Weber, H., T. Polen, J. Heuveling, V. Wendisch, and R. Hengge (2005) Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters and sigma factor selectivity. J. Bacteriol. 187, 1591-1603.
Three different growth and stress conditions that are known to result in high cellular σS levels were used for RNA preparation:
- growth in LB medium to an OD578 of 4.0 (this corresponds to transition into stationary phase);
- growth in minimal medium to which 0.3 M NaCl was added at an OD578 of 0.3 (cells were harvested 20 min after this hyperosmotic shift);
- growth in LB medium which was acidified by the addition of MES at an OD578 of 0.4 (cells were harvested 40 min after this shift to pH5).
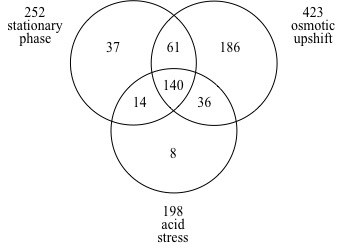
We identified a total of 481 genes, which exhibited more than two-fold higher expression in the rpoS+ strain than in the rpoS mutant (Fig. 1). Only 140 of these positively σS-controlled genes were found under all three growth and stress conditions (Fig. 1). These genes are refer to as the “core” set of σS-controlled genes. The other 341 genes revealed their σS-dependence only under one or two of the growth and stress conditions used (with genes in all possible combinatorial groups; Fig. 1). The table shows expression ratios for rpoS+ and rpoS- strains of all σS-controlled genes (not only “core“ genes).
| gene | bnum | OD 4.0 | NaCl | pH 5 | gene product |
| acrD | b2470 | 1.596 | 3.645 | 1.835 | aminoglycoside/multidrug efflux pump (RND family) |
| adhP | b1478 | 5.379 | 9.190 | 3.425 | alcohol dehydrogenase, propanol-preferring |
| agp | b1002 | 0.947 | 4.191 | 1.536 | glucose-1-phosphatase, also has inositol phosphatase activity |
| aidB | b4187 | 4.105 | 2.080 | 3.467 | putative acyl-CoA dehydrogenase (flavoprotein) , adaptive response (transcription activated by Ada) |
| aldB | b3588 | 4.348 | 2.590 | 1.563 | aldehyde dehydrogenase B (lactaldehyde dehydrogenase) |
| amyA | b1927 | 12.209 | 11.015 | 10.568 | cytoplasmic alpha-amylase |
| appC | b0978 | 3.291 | 1.690 | 0.890 | cytochrome oxidase bd-II, subunit I |
| appY | b0564 | 2.094 | 0.879 | 1.954 | DLP12 prophage; transcriptional regulator required for anaerobic and stationary phase induction of genes (AraC/XylS family) |
| arcA | b4401 | 2.737 | 2.687 | 1.370 | response regulator in two-component regulatory system with ArcB (or CpxA), regulates respiratory and fermentative metabolism (OmpR family) |
| argG | b3172 | 1.991 | 2.117 | 1.858 | argininosuccinate synthetase |
| argH | b3960 | 1.441 | 2.346 | 1.147 | argininosuccinate lyase |
| arnB | b2253 | 2.729 | 9.043 | 1.548 | uridine 5'-(beta-1-threo-pentapyranosyl-4-ulose diphosphate) aminotransferase, PLP dependent |
| aroM | b0390 | 2.470 | 3.659 | 1.638 | protein of aro operon, regulated by aroR |
| artJ | b0860 | 1.941 | 0.897 | 2.337 | arginine transport protein (ABC superfamily, peri_bind) |
| artM | b0861 | 2.196 | 2.788 | 2.606 | arginine transport protein (ABC superfamily, membrane) |
| artP | b0864 | 3.497 | 4.557 | 4.710 | arginine transport protein (ABC superfamily, atp_bind) |
| artQ | b0862 | 2.107 | 2.417 | 2.065 | arginine transport protein (ABC superfamily, membrane) |
| b0235 | b0235 | 1.592 | 2.042 | 1.155 | unknown CDS |
| b0609 | b0609 | 0.892 | 3.670 | 0.766 | unknown CDS |
| b0795 | b0795 | 1.252 | 2.761 | 0.986 | putative multidrug resistance membrane protein |
| b1173 | b1173 | 1.156 | 2.272 | 1.341 | unknown CDS |
| b1341 | b1341 | 6.740 | 9.543 | 3.148 | conserved protein with PYP-like sensor domain |
| b1342 | b1342 | 1.825 | 2.840 | 1.678 | putative membrane associated protein |
| b1428 | b1428 | 0.000 | 8.318 | 7.804 | putative LpxA-like enzyme |
| b1454 | b1454 | 3.415 | 13.772 | 3.643 | putative glutathione S-transferase with thioredoxin-like and glutathione S-transferases, C-terminal domain |
| b1520 | b1520 | 2.366 | 3.046 | 1.473 | conserved hypothetical protein |
| b1547 | b1547 | 2.254 | 4.706 | 2.350 | Qin prophage; putative tail fiber protein |
| b1598 | b1598 | 2.606 | 5.240 | 1.188 | putative enzyme with serine protease-like domain |
| b1664 | b1664 | 2.332 | 2.799 | 1.058 | putative enzyme with pectin lyase-like domain |
| b1668 | b1668 | 2.138 | 2.341 | 2.610 | putative oxidoreductase with NAD(P)/FAD-binding domain |
| b1672 | b1672 | 1.820 | 3.020 | 1.859 | conserved hypothetical protein |
| b1674 | b1674 | 1.299 | 2.696 | 1.573 | putative 4Fe-4S ferredoxin-type protein |
| b1806 | b1806 | 2.434 | 3.209 | 4.055 | putative membrane protein |
| b1810 | b1810 | 3.020 | 3.334 | 2.720 | conserved hypothetical protein |
| b1836 | b1836 | 3.784 | 2.493 | 2.659 | unknown CDS |
| b1956 | b1956 | 2.417 | 3.635 | 1.878 | putative PLP-dependent transferase |
| b1957 | b1957 | 5.462 | 3.018 | 2.146 | unknown CDS |
| b2475 | b2475 | 1.879 | 2.109 | 1.858 | conserved protein |
| b2858 | b2858 | 2.610 | 3.078 | 3.008 | unknown CDS |
| b3000 | b3000 | 2.514 | 3.790 | 1.683 | putative enzyme with alpha/beta-hydrolase domain |
| b3023 | b3023 | 2.636 | 2.506 | 1.445 | conserved protein with probable bacterial effector-binding domain |
| basR | b4113 | 1.685 | 2.093 | 1.218 | response regulator in two-component regulatory system with BasS (OmpR family) |
| basS | b4112 | 2.408 | 1.448 | 1.487 | sensory histidine kinase in two-component regulatory system with BasR |
| bfr | b3336 | 8.844 | 3.283 | 4.717 | bacterioferritin, an iron storage homoprotein |
| blc | b4149 | 4.772 | 8.988 | 8.097 | outer membrane lipoprotein (lipocalin) |
| bolA | b0435 | 4.742 | 8.472 | 4.768 | transcriptional activator of morphogenic pathway (BolA family), important in general stress response |
| btuD | b1709 | 1.885 | 8.369 | 1.034 | vitamin B12 transport protein (ABC superfamily, atp_bind) |
| btuE | b1710 | 2.543 | 7.261 | 1.177 | vitamin B12 transport protein (ABC superfamily, peri_bind) |
| cbpA | b1000 | 4.970 | 4.159 | 2.045 | curved DNA-binding protein, co-chaperone of DnaK (Hsp40 family) |
| cedA | b1731 | 0.000 | 2.045 | 0.000 | cell division activator |
| cfa | b1661 | 4.842 | 3.407 | 1.457 | cyclopropane fatty acyl phospholipid synthase (unsaturated-phospholipid methyltransferase) |
| chaB | b1217 | 3.357 | 5.542 | 3.383 | cation transport regulator |
| chaC | b1218 | 1.575 | 3.071 | 1.316 | cation transport regulator |
| cheA | b1888 | 2.112 | 4.182 | 2.391 | multimodular: chemotactic sensory histidine kinase (soluble) in two-component regulatory system with CheB and CheY, senses chemotactic signal |
| cheY | b1882 | 1.607 | 3.037 | 1.236 | chemotactic response regulator in two-component regulatory system with CheA, transmits signals to FliM flagelllar motor component |
| cheZ | b1881 | 1.926 | 3.491 | 1.706 | chemotactic response, CheY protein phophatase |
| chiA | b3338 | 2.046 | 2.366 | 1.601 | endochitinase, periplasmic |
| clpX | b0438 | 1.561 | 2.145 | 1.210 | ATPase, chaperone subunit of serine protease |
| cpxP | b3913 | 0.752 | 1.061 | 2.046 | periplasmic repressor of cpx regulon by interaction with CpxA |
| cpxR | b3912 | 1.785 | 2.088 | 1.356 | response regulator in two-component regulatory system with CpxA, regulates genes involved in folding/degrading periplasmic proteins (OmpR family) |
| csgB | b1041 | 1.570 | 2.620 | 1.472 | minor curlin subunit precursor, nucleator for assembly of adhesive surface organelles |
| csiD | b2659 | 6.902 | 7.822 | 3.684 | conserved protein with clavaminate synthase-like domain |
| csiE | b2535 | 1.463 | 2.847 | 1.868 | stationary phase-inducible protein with PTS-regulatory domain |
| cueR | b0487 | 2.502 | 1.728 | 1.561 | transcriptional repressor for CopA a copper exporting ATPase (MerR family) |
| cutA | b4137 | 2.012 | 0.763 | 0.876 | periplasmic divalent cation tolerance protein; cytochrome c biogenesis |
| cybC | b4236 | 1.755 | 2.034 | 1.213 | cytochrome b(562) |
| cysH | b2762 | 2.205 | 0.566 | 0.153 | 3'-phosphoadenosine 5'-phosphosulfate (PAPS) reductase |
| cysQ | b4214 | 1.612 | 4.319 | 1.876 | protein that acts on 3'-phosphoadenosine-5'-phosphosulfate with sugar phosphatase domain |
| dacC | b0839 | 1.653 | 3.822 | 1.832 | D-alanyl-D-alanine carboxypeptidase; penicillin-binding protein 6a |
| dbpA | b1343 | 2.547 | 3.846 | 1.241 | ATP-dependent RNA helicase, stimulated by 23S rRNA |
| ddpA | b1487 | 2.145 | 0.911 | 1.459 | putative dipeptide transport protein (ABC superfamily, peri_bind) |
| ddpF | b1483 | 2.008 | 5.462 | 1.458 | putative dipeptide transport protein (ABC superfamily, atp_bind) |
| deoA | b4382 | 1.455 | 2.151 | 1.576 | thymidine phosphorylase |
| deoC | b4381 | 1.960 | 2.351 | 1.428 | 2-deoxyribose-5-phosphate aldolase, NAD(P)-linked |
| deoD | b4384 | 1.732 | 2.284 | 1.443 | purine-nucleoside phosphorylase |
| dkgV | b3012 | 4.365 | 6.334 | 2.366 | 2,5-diketo-D-gluconate reductase A |
| dppA | b3544 | 3.457 | 0.481 | 1.321 | dipeptide transport protein (ABC superfamily, peri_bind) |
| dppB | b3543 | 2.624 | 0.394 | 1.146 | dipeptide transport protein 1 (ABC superfamily, membrane) |
| dppC | b3542 | 2.438 | 0.496 | 1.180 | dipeptide transport protein 2 (ABC superfamily, membrane) |
| dppD | b3541 | 2.510 | 0.476 | 1.047 | dipeptide transport protein (ABC superfamily, atp_bind) |
| dppF | b3540 | 2.714 | 0.552 | 1.347 | dipeptide transport protein (ABC superfamily, atp_bind) |
| dps | b0812 | 8.919 | 24.192 | 4.319 | stress response DNA-binding protein with ferritin-like domain |
| dsbG | b0604 | 2.594 | 2.855 | 1.968 | disulfide isomerase, thiol-disulphide oxidase, periplasmic |
| dsrB | b1952 | 2.404 | 2.258 | 1.554 | conserved hypothetical protein; regulated by DsrA and HNS, under control of RpoS |
| elaB | b2266 | 8.110 | 17.824 | 11.803 | unknown CDS |
| erfK | b1990 | 2.239 | 4.550 | 2.176 | conserved hypothetical protein with NAD(P)-binding Rossmann-fold domain |
| eutH | b2452 | 3.105 | 3.057 | 2.594 | putative transport protein, ethanolamine utilization |
| exuT | b3093 | 0.923 | 2.291 | 1.135 | hexuronate transport protein (MFS family) |
| fbaB | b2097 | 7.998 | 9.609 | 9.325 | fructose-bisphosphate aldolase class I |
| fhlA | b2731 | 2.751 | 4.536 | 1.849 | transcriptional activator for induction of formate hydrogen-lyase (EBP family) |
| fic | b3361 | 12.942 | 17.756 | 10.823 | possible cell filamentation protein, induced in stationary phase |
| flgC | b1074 | 1.500 | 2.535 | 1.572 | flagellar biosynthesis; cell-proximal portion of basal-body rod |
| flhE | b1878 | 2.232 | 1.769 | 1.587 | flagellar protein |
| fliJ | b1942 | 2.011 | 1.026 | 1.242 | flagellar fliJ protein |
| flxA | b1566 | 2.646 | 2.360 | 1.112 | Qin prophage |
| focA | b0904 | 1.454 | 6.314 | 1.858 | formate transport protein (formate channel 1) (FNT family) |
| frdA | b4154 | 1.025 | 2.997 | 1.146 | fumarate reductase, anaerobic, catalytic and NAD/flavoprotein subunit |
| fruA | b2167 | 2.307 | 2.682 | 1.971 | multimodular: PTS family enzyme IIB'B (N-terminal); enzyme IIC (C-terminal), fructose-specific |
| fruK | b2168 | 0.000 | 4.140 | 1.866 | fructose-1-phosphate kinase |
| fsa | b0825 | 0.000 | 4.453 | 1.050 | fructose-6-phosphate aldolase 1 |
| fucA | b2800 | 1.064 | 2.415 | 1.392 | L-fuculose-1-phosphate aldolase |
| gabD | b2661 | 4.805 | 4.077 | 2.886 | succinate-semialdehyde dehydrogenase I, NADP-dependent |
| gabP | b2663 | 4.064 | 4.703 | 2.828 | gamma-aminobutyrate transport protein, RpoS-dependent (APC family) |
| gabT | b2662 | 2.088 | 2.042 | 1.574 | 4-aminobutyrate aminotransferase, PLP-dependent |
| gadA | b3517 | 32.384 | 5.803 | 2.332 | glutamate decarboxylase A, isozyme, PLP-dependent |
| gadB | b1493 | 16.444 | 3.963 | 2.220 | glutamate decarboxylase, PLP-dependent, isozyme beta |
| gadC | b1492 | 10.280 | 3.478 | 2.223 | putative glutamate:gamma-aminobutyric acid antiporter (APC family) |
| gadE | b3512 | 13.062 | 8.680 | 1.142 | transcriptional regulator for gadABC operon, activates glutamate decarhboxylase-dependent acid resistance |
| gadW | b3515 | 6.119 | 5.975 | 3.093 | transcriptional regulator for GadX (regulatory protein), glutamic acid decarboxylase (GadA,B), and glutamate transport protein (GadC) (AraC/XylS family) |
| gadX | b3516 | 6.091 | 7.447 | 3.138 | transcriptional regulator for glutamic acid decarboylase and transporter (gadA, gad BC) (AraC/XylS family) |
| gapC_1 | b1417 | 1.505 | 2.449 | 1.817 | split gene, glyceraldehyde-3-phosphate dehydrogenase C (first fragment of interrupted enzyme gene) |
| gapC_2 | b1416 | 0.783 | 2.171 | 1.463 | split gene, glyceraldehyde-3-phosphate dehydrogenase C (second fragment of interrupted enzyme gene) |
| garK | b3124 | 2.102 | 2.988 | 1.705 | glycerate kinase I |
| garL | b3126 | 1.816 | 2.924 | 1.922 | alpha-dehydro-beta-deoxy-D-glucarate aldolase |
| gcd | b0124 | 1.199 | 1.153 | 2.097 | glucose dehydrogenase |
| gdhA | b1761 | 2.045 | 1.461 | 3.064 | glutamate dehydrogenase, NADP-specific |
| gem | b1285 | 6.782 | 3.825 | 6.942 | RNase II modulator with PYP-like sensor domain |
| ggt | b3447 | 4.656 | 5.188 | 2.015 | gamma-glutamyltranspeptidase |
| glcA | b2975 | 1.597 | 2.004 | 0.701 | glycolate permease (LctP family) |
| glcB | b2976 | 1.341 | 2.972 | 1.589 | malate synthase G |
| glgS | b3049 | 1.311 | 3.929 | 2.466 | glycogen biosynthesis, rpoS-dependent |
| gloA | b1651 | 1.510 | 2.174 | 1.378 | glyoxalase I, nickel isomerase |
| hcr | b0872 | 2.489 | 2.983 | 1.959 | NADH oxidoreductase for hcp gene product |
| hdeA | b3510 | 11.263 | 9.162 | 1.646 | conserved protein with protein HNS-dependent expression A; HdeA-like domain |
| hdeB | b3509 | 6.607 | 4.463 | 1.281 | conserved hypothetical protein |
| hdeD | b3511 | 2.522 | 2.070 | 1.265 | putative membrane protein |
| hdhA | b1619 | 2.710 | 6.972 | 2.428 | 7alpha-hydroxysteroid dehydrogenase, NAD-dependent |
| hemH | b0475 | 1.541 | 3.105 | 1.515 | ferrochelatase |
| himA | b1712 | 1.753 | 2.598 | 1.393 | integration host factor (IHF), alpha subunit, DNA-bending protein, DNA replication |
| hlyE | b1182 | 2.793 | 1.499 | 0.000 | hemolysin E, pore-forming cytotoxin |
| hnr | b1235 | 4.677 | 7.194 | 4.077 | response regulator involved in protein turnover, controls stability of RpoS |
| hscA | b2526 | 2.105 | 5.491 | 0.934 | chaperone (Hsp70 family), believed to be involved in assembly of Fe-S clusters |
| hslO | b3401 | 1.956 | 2.535 | 1.395 | heat shock protein 33, redox regulated chaperone |
| hycF | b2720 | 10.674 | 24.062 | 9.113 | hydrogenase 3, Fe-S related |
| hycI | b2717 | 3.996 | 4.162 | 2.793 | protease involved in processing C-terminal end of HycE |
| hyfD | b2484 | 1.579 | 2.746 | 1.575 | hydrogenase 4, D membrane subunit |
| idi | b2889 | 1.494 | 2.549 | 1.538 | isopentenyl diphosphate isomerase |
| ivbL | b3672 | 2.026 | 1.422 | 1.116 | ilvB operon leader peptide |
| ivy | b0220 | 2.242 | 1.539 | 1.135 | inhibitor of vertebrate C-type lysozyme, periplasmic |
| katE | b1732 | 11.143 | 8.995 | 7.368 | catalase; hydroperoxidase HPII (III), RpoS-dependent |
| kbl | b3617 | 0.712 | 2.287 | 0.908 | 2-amino-3-ketobutyrate CoA ligase (glycine acetyltransferase) |
| kch | b1250 | 2.549 | 1.632 | 1.098 | putative potassium channel protein with NAD(P)-binding Rossmann-fold domain (VIC family) |
| lasT | b4403 | 1.147 | 2.145 | 1.103 | putative tRNA/rRNA methyltransferase |
| ldcC | b0186 | 3.758 | 3.718 | 3.879 | lysine decarboxylase 2, constitutive |
| ltaA | b0870 | 1.127 | 3.365 | 2.040 | L-allo-threonine aldolase, PLP-dependent |
| luxS | b2687 | 1.959 | 2.412 | 1.478 | quorum-sensing protein, produces autoinducer - acyl-homoserine lactone-signaling molecules |
| mak | b0394 | 1.543 | 2.669 | 1.470 | manno(fructo) kinase |
| malP | b3417 | 1.479 | 8.703 | 5.224 | maltodextrin phosphorylase |
| malQ | b3416 | 1.243 | 3.090 | 1.905 | 4-alpha-glucanotransferase (amylomaltase) |
| manX | b1817 | 1.050 | 2.342 | 0.817 | multimodular: PTS family enzyme IIA (N-terminal); enzyme IIB (C-terminal), mannose-specific |
| marC | b1529 | 0.000 | 2.543 | 1.862 | putative MarC transport protein, multiple antibiotic resistance protein |
| menD | b2264 | 1.383 | 2.293 | 1.687 | bifunctional: 2-oxoglutarate decarboxylase; SHCHC synthase |
| mlrA | b2127 | 7.079 | 3.018 | 5.715 | transcriptional regulator of curli and extracellular matrix synthesis (MerR family) |
| mltB | b2701 | 1.791 | 2.938 | 1.459 | membrane-bound lytic murein transglycosylase B |
| mntH | b2392 | 2.174 | 2.384 | 1.718 | manganese/divalent cation transport protein (NRAMP family) |
| mrcA | b3396 | 1.414 | 2.547 | 0.923 | multimodular penicillin-binding protein 1a: transglycosylase (N-terminal); transpeptidase (C-terminal) |
| mscL | b3291 | 2.940 | 2.314 | 2.586 | mechanosensitive channel |
| mscS | b2924 | 2.729 | 3.276 | 1.067 | mechanosensitive channel protein (MscS family) |
| msyB | b1051 | 17.365 | 8.597 | 8.885 | acidic protein suppresses mutants lacking function of protein export |
| mtlA | b3599 | 1.042 | 2.203 | 1.492 | multimodular: PTS family enzyme IICB (N-terminal); enzyme IIA (C-terminal), mannitol-specific |
| mug | b3068 | 1.835 | 2.531 | 2.032 | DNA glycosylase, G/U mismatch-specific |
| nagB | b0678 | 1.986 | 3.324 | 1.951 | glucosamine-6-phosphate deaminase |
| nanK | b3222 | 1.926 | 11.203 | 1.340 | putative ManNAc kinase with Actin-like ATPase domain/transcriptional regulator (NagC/XylR (ROK) family) |
| napH | b2204 | 0.938 | 2.070 | 0.832 | ferredoxin-type protein, electron transfer |
| narI | b1227 | 2.716 | 2.234 | 1.318 | nitrate reductase 1, cytochrome b(NR), gamma subunit |
| narU | b1469 | 2.750 | 3.617 | 2.067 | nitrate extrusion protein (MFS family) |
| narY | b1467 | 5.244 | 5.697 | 4.567 | nitrate reductase 2, beta subunit |
| nfi | b3998 | yjaF | 0.998 | 2.203 | endonuclease V (deoxyinosine 3'endoduclease) |
| nhaA | b0019 | 3.744 | 3.905 | 1.641 | sodium:proton antiporter (NhaA family) |
| nifJ | b1378 | 1.605 | 2.138 | 1.407 | multimodular: putative pyruvate-flavodoxin oxidoreductase, Fe-S subunit |
| nlpC | b1708 | 1.026 | 2.149 | 0.867 | lipoprotein |
| norV | b2710 | 1.361 | 2.270 | 0.963 | flavorubredoxin (FIRd), bifunctional NO and O2 reductase |
| nrdD | b4238 | 1.705 | 10.178 | 2.117 | anaerobic ribonucleoside-triphosphate reductase |
| nrdE | b2675 | 3.562 | 1.164 | 3.840 | ribonucleoside-diphosphate reductase 2, alpha subunit |
| ompA | b0957 | 1.950 | 2.386 | 1.312 | outer membrane protein 3a (II*;G;d) |
| ompG | b1319 | 0.000 | 6.060 | 0.000 | outer membrane pore protein |
| ompR | b3405 | 1.321 | 2.086 | 1.204 | response regulator in two-component regulatory system with EnvZ, regulates ompF and ompC expression (OmpR family) |
| ompX | b0814 | 2.153 | 2.516 | 1.112 | outer membrane protease, receptor for phage OX2 |
| osmB | b1283 | 3.342 | 2.360 | 2.337 | lipoprotein, osmotically inducible |
| osmC | b1482 | 4.932 | 6.185 | 3.513 | resistance protein, osmotically inducible |
| osmY | b4376 | 50.660 | 68.760 | 19.084 | hyperosmotically inducible periplasmic protein, RpoS-dependent stationary phase gene |
| otsA | b1896 | 10.268 | 52.440 | 13.793 | trehalose-6-phosphate synthase |
| otsB | b1897 | 5.555 | 38.224 | 9.085 | trehalose-6-phosphate phosphatase, biosynthetic |
| paaX | b1399 | 1.182 | 2.189 | 1.772 | transcriptional repressor for phenylacetic acid degradation |
| paaY | b1400 | 1.146 | 3.196 | 2.153 | putative acyl transferase with trimeric LpxA-like domain , iron-binding |
| pabA | b3360 | 1.552 | 3.043 | 2.363 | aminodeoxychorismate synthase subunit II, component of p-aminobenzoate synthase multienzyme complex |
| pdhR | b0113 | 5.008 | 3.417 | 2.855 | transcriptional repressor for pyruvate dehydrogenase complex (GntR family) |
| pfkA | b3916 | 1.501 | 1.947 | 2.084 | 6-phosphofructokinase I |
| pfkB | b1723 | 2.072 | 2.489 | 1.709 | 6-phosphofructokinase II |
| phnA | b4108 | 1.204 | 0.988 | 2.196 | putative alkylphosphonate uptake protein in phosphonate metabolism |
| phnB | b4107 | 4.805 | 5.366 | 3.381 | conserved protein with glyoxalase and dihydroxybiphenyl dioxygenase domain |
| phnO | b4093 | 1.488 | 2.477 | 1.319 | putative acyl-CoA N-acyltransferase |
| phrB | b0708 | 1.980 | 6.437 | 4.550 | deoxyribodipyrimidine photolyase (photoreactivation), FAD-binding |
| pinQ | b1545 | 1.262 | 2.253 | 0.000 | Qin prophage; putative resolvase (recombinase) |
| pinR | b1374 | 1.383 | 2.021 | 0.971 | Rac prophage; putative transposon resolvase |
| pitA | b3493 | 1.104 | 2.163 | 1.727 | low-affinity phosphate transport protein (PiT family) |
| polB | b0060 | 1.956 | 3.570 | 1.609 | DNA polymerase II and 3' -> 5' exonuclease |
| potF | b0854 | 2.756 | 4.246 | 2.244 | putrescine transport protein (ABC superfamily, peri_bind) |
| potG | b0855 | 1.941 | 2.763 | 1.274 | putrescine transport protein (ABC superfamily, atp_bind) |
| poxA | b4155 | 2.159 | 2.624 | 1.782 | putative lysyl-tRNA synthetase with Class II aaRS and biotin synthetase domains |
| poxB | b0871 | 17.674 | 45.885 | 29.603 | pyruvate dehydrogenase/oxidase: FAD- and thiamin PPi-binding |
| pphA | b1838 | 1.484 | 2.291 | 1.336 | serine/threonine-specific protein phosphatase 1, signals protein misfolding |
| prlC | b3498 | 1.652 | 2.200 | 1.402 | oligopeptidase A |
| proP | b4111 | 4.903 | 0.794 | 1.401 | proline transport protein, low-affinity (proline permease II) (MFS family) |
| psiF | b0384 | 9.477 | 10.528 | 7.419 | induced by phosphate starvation |
| ptrB | b1845 | 1.514 | 2.929 | 2.026 | protease II |
| ptsG | b1101 | 2.251 | 2.393 | 1.749 | multimodular: PTS family enzyme IIC (N-terminal); enzyme IIB (C-terminal), glucose-specific |
| pykA | b1854 | 0.886 | 2.561 | 0.826 | pyruvate kinase II, glucose-stimulated |
| qor | b4051 | 2.390 | 2.731 | 2.638 | quinone oxidoreductase, NADPH-dependent |
| rbn | b3886 | yihY | 1.005 | 2.003 | tRNA processing exoribonuclease BN |
| rfaS | b3629 | 2.713 | 2.246 | 1.349 | lipopolysaccharide core biosynthesis |
| rhlB | b3780 | 2.574 | 2.303 | 1.710 | putative ATP-dependent helicase with nucleoside triP hydrolase domain |
| rhsC | b0701 | 0.000 | 3.076 | 1.472 | part of RhsC protein (b0700) in RhsC element |
| rluA | b0058 | 1.839 | 2.428 | 1.827 | 23S rRNA pseudouridylate 746 synthase |
| rluD | b2594 | 1.639 | 2.588 | 1.389 | pseudouridine synthase (pseudouridines 1911, 1915, 1917 in 23S RNA) |
| rluE | b1135 | 1.728 | 2.050 | 1.310 | pseudouridine synthase (makes pseudouridine2457 in 23 S RNA) |
| rnk | b0610 | 0.882 | 2.430 | 0.653 | regulator of nucleoside diphosphate kinase |
| rpsV | b1480 | 5.204 | 6.072 | 4.599 | 30S ribosomal subunit protein S22 |
| rtcB | b3421 | 1.253 | 2.116 | 1.484 | putative PLP-dependent transferase |
| sbmC | b2009 | 1.721 | 3.093 | 1.501 | DNA gyrase inhibitor |
| sfsA | b0146 | 1.277 | 3.043 | 1.405 | transcriptional regulator of maltose metabolism |
| slp | b3506 | 3.681 | 2.790 | 1.077 | outer membrane protein, induced after carbon starvation |
| sodC | b1646 | 1.878 | 3.828 | 2.234 | superoxide dismutase precursor (Cu-Zn) |
| speB | b2937 | 1.256 | 1.709 | 2.342 | agmatinase |
| ssnA | b2879 | 1.452 | 5.580 | 0.000 | putative protein with metallo-dependent hydrolase domain, commonly a deaminase |
| ssuE | b0937 | 6.095 | 1.602 | 3.416 | NAD(P)H-dependent FMN reductase, sulfate starvation-induced protein |
| sufA | b1684 | 10.241 | 1.140 | 3.086 | Fe-S cluster assembly protein |
| sufB | b1683 | 13.762 | 1.064 | 3.510 | putative transport protein associated with Fe-S cluster assembly |
| sufC | b1682 | 4.178 | 1.031 | 3.214 | putative transport protein associated with Fe-S cluster assembly (ABC superfamily, atp_bind) |
| sufD | b1681 | 6.233 | 0.975 | 3.864 | required for stability of Fe-S component of FhuF |
| sufE | b1679 | 3.381 | 1.019 | 3.420 | stimulator of SufS activity |
| sufS | b1680 | 4.761 | 1.081 | 4.058 | selenocysteine lyase, PLP-dependent |
| sugE | b4148 | 1.917 | 2.620 | 2.754 | quarternary ammonium compound transport protein (SMR family) |
| talA | b2464 | 6.792 | 13.082 | 13.825 | transaldolase A |
| tam | b1519 | 2.758 | 8.683 | 3.532 | trans-aconitate methyltransferase |
| tdh | b3616 | 0.747 | 2.547 | 0.968 | threonine 3-dehydrogenase, NAD(P)-binding |
| tesB | b0452 | 1.379 | 2.201 | 1.144 | acyl-CoA thioesterase II |
| thiG | b3991 | 2.201 | 1.581 | 1.124 | thiamin biosynthesis enzyme subunit, with ThiH |
| tktB | b2465 | 14.077 | 35.865 | 10.756 | transketolase 2, thiamin-binding, isozyme |
| treA | b1197 | 7.980 | 12.706 | 2.502 | trehalase, periplasmic |
| treF | b3519 | 2.070 | 9.254 | 5.204 | trehalase, cytoplasmic |
| trmA | b3965 | 1.639 | 2.127 | 1.383 | tRNA (uracil-5-)-methyltransferase |
| trpH | b1266 | 1.792 | 2.253 | 1.501 | conserved hypothetical protein |
| udk | b2066 | 1.960 | 3.664 | 1.718 | uridine/cytidine kinase |
| ugpB | b3453 | 3.294 | 5.546 | 3.289 | sn-glycerol 3-phosphate transport protein (ABC superfamily, peri_bind) |
| ugpC | b3450 | 2.391 | 2.500 | 2.443 | sn-glycerol 3-phosphate transport protein (ABC superfamily, atp_bind) |
| ugpQ | b3449 | 2.298 | 3.687 | 2.008 | glycerophosphodiester phosphodiesterase, cytosolic |
| uspB | b3494 | 4.522 | 5.907 | 3.609 | universal stress protein B |
| wrbA | b1004 | 9.376 | 6.834 | 14.768 | flavodoxin-like protein, trp repressor-binding protein |
| yaaJ | b0007 | 1.406 | 3.996 | 0.861 | putative alanine/glycine transport protein (AGCS family) |
| yadQ | b0155 | 1.148 | 2.067 | 0.863 | putative chlorine transport protein (ClC family) |
| yaeJ | b0191 | 1.530 | 2.171 | 1.203 | conserved protein with RF2 (polypeptide chain release factor 2) domain |
| yaeR | b0187 | 1.340 | 2.149 | 1.679 | putative enzyme with glyoxalase and dihydroxybiphenyl dioxygenase domains |
| yafV | b0219 | 1.181 | 2.612 | 1.471 | putative NAD(P)-binding amidase-type enzyme with nitrilase/N-carbamoyl-D-aminoacid amidohydrolase |
| yaiA | b0389 | 3.917 | 4.724 | 2.410 | unknown CDS |
| yajO | b0419 | 2.211 | 6.761 | 3.422 | putative oxidoreductase, NAD(P)-dependent |
| ybaA | b0456 | 4.375 | 4.617 | 1.531 | conserved hypothetical protein |
| ybaL | b0478 | 1.596 | 2.805 | 1.413 | putative transport protein with NAD(P)-binding Rossmann-fold domain (CPA2 family) |
| ybaS | b0485 | 14.050 | 9.462 | 2.698 | putative glutaminase/carboxypeptidase with beta-Lactamase/D-ala carboxypeptidase domain |
| ybaW | b0443 | 2.042 | 3.301 | 1.135 | conserved protein with thioesterase/thiol ester dehydrase-isomerase domain |
| ybaY | b0453 | 6.147 | 8.670 | 6.086 | glycoprotein/polysaccharide metabolism |
| ybaZ | b0454 | 1.121 | 2.059 | 1.166 | putative methylated DNA-protein cysteine methyltransferase, C-terminal domain |
| ybdK | b0581 | 8.777 | 14.701 | 12.706 | conserved hypothetical protein |
| ybdR | b0608 | 0.000 | 10.343 | 1.218 | putative dehydrogenase with NAD(P)-binding and GroES domains |
| ybeL | b0643 | 2.415 | 9.802 | 4.266 | conserved hypothetical protein |
| ybeM | b0626 | 2.104 | 2.265 | 1.900 | putative NAD(P)-binding amidohydrolase (C-N hydrolase family) |
| ybeV | b0649 | 0.951 | 2.246 | 0.786 | putative chaperone with dnaJ-like domain |
| ybgA | b0707 | 6.531 | 16.969 | 13.243 | conserved protein |
| ybgS | b0753 | 8.427 | 38.077 | 16.119 | conserved protein |
| ybhB | b0773 | 2.314 | 2.737 | 1.799 | conserved protein with phosphatidylethanolamine-binding domain |
| ybhE | b0767 | 2.659 | 5.572 | 2.375 | putative isomerase |
| ybhF | b0794 | 1.626 | 2.151 | 0.982 | multimodular: putative transport protein (ABC superfamily, atp_bind) |
| ybhN | b0788 | 0.000 | 3.355 | 0.000 | putative negative regulator |
| ybhO | b0789 | 5.861 | 12.199 | 10.576 | cardiolipin (CL) synthase 2 |
| ybhR | b0792 | 3.013 | 7.986 | 10.471 | putative transport protein (ABC superfamily, membrane) |
| ybiH | b0796 | 0.000 | 3.625 | 1.059 | putative transcriptional repressor with homeodomain-like DNA binding domain (TetR/AcrR family) |
| ybiI | b0803 | 0.000 | 4.046 | 4.845 | conserved hypothetical protein |
| ybiM | b0806 | 2.267 | 2.671 | 1.861 | conserved hypothetical protein |
| ybiN | b0807 | 1.567 | 3.124 | 1.897 | putative methyltransferase with S-adenosyl-L-methionine-dependent methyltransferase domain |
| ybiO | b0808 | 8.492 | 4.512 | 2.021 | putative transport protein, integral membrane location |
| ybiP | b0815 | 7.727 | 5.888 | 2.427 | putative transmembrane protein (N-terminal); putative phosphatase (C-terminal) |
| ybjP | b0865 | 3.659 | 6.531 | 6.242 | conserved hypothetical protein |
| ybjQ | b0866 | 1.977 | 3.055 | 1.469 | conserved hypothetical protein |
| ybjR | b0867 | 1.476 | 2.563 | 1.561 | putative amidase |
| ybjT | b0869 | 0.933 | 2.032 | 1.555 | conserved protein with ATP-binding and NAD(P)-binding domains |
| ycaC | b0897 | 13.335 | 5.916 | 5.875 | putative enzyme with cysteine hydrolase domain |
| ycaP | b0906 | 4.295 | 7.586 | 4.617 | conserved hypothetical protein |
| ycbB | b0925 | 2.138 | 1.956 | 1.903 | putative carboxypeptidasewith PGDB-like domain |
| ycbK | b0926 | 1.642 | 2.282 | 1.305 | conserved hypothetical protein with Hedgehog/DD-pepidase domain |
| yccE | b1001 | 2.861 | 2.907 | 1.797 | putative hemoglobin-binding protein |
| yccJ | b1003 | 10.423 | 17.087 | 16.331 | unknown CDS |
| yccU | b0965 | 0.000 | 2.578 | 2.054 | putative NAD(P)-binding enzyme |
| ycdF | b1005 | 7.396 | 15.584 | 19.800 | unknown CDS |
| ycdG | b1006 | 3.184 | 2.572 | 1.567 | putative uracil transport protein (NCS2 family) |
| yceK | b1050 | 14.928 | 14.499 | 10.139 | unknown CDS |
| ycfF | b1103 | 2.677 | 2.988 | 2.104 | putative inhibitor of protein kinase C, contains a transferase domain |
| ycfH | b1100 | 2.390 | 2.502 | 1.709 | putative metallo-dependent hydrolase (with domain) |
| ycfM | b1105 | 1.803 | 2.386 | 1.430 | putative fibronectin-binding protein |
| ycfR | b1112 | 1.656 | 2.240 | 1.842 | conserved hypothetical protein |
| ycgB | b1188 | 10.359 | 35.427 | 11.194 | conserved hypothetical protein |
| ycgG | b1168 | 1.359 | 2.460 | 1.554 | conserved protein |
| ycgZ | b1164 | 5.109 | 5.525 | 3.999 | unknown CDS |
| ychH | b1205 | 0.966 | 3.617 | 0.000 | conserved hypothetical protein |
| ychK | b1234 | 2.500 | 3.189 | 2.261 | putative transmembrane protein |
| yciE | b1257 | 1.895 | 16.660 | 0.000 | conserved protein with ferritin-like domain |
| yciF | b1258 | 2.981 | 20.433 | 2.039 | conserved protein |
| yciG | b1259 | 18.155 | 84.918 | 19.187 | conserved hypothetical protein |
| yciQ | b1268 | 1.392 | 2.150 | 0.000 | putative membrane protein |
| ycjY | b1327 | 0.000 | 2.205 | 0.000 | conserved hypothetical protein, alpha/beta-hydrolase domain |
| ydbC | b1406 | 1.130 | 2.009 | 0.910 | putative oxidoreductase, NAD(P)-binding domain |
| ydcD | b1457 | 2.765 | 1.049 | 1.075 | unknown CDS |
| ydcJ | b1423 | 7.362 | 1.738 | 1.324 | conserved hypothetical protein |
| ydcS | b1440 | 21.660 | 4.831 | 4.362 | putative transport protein (ABC superfamily, peri_bind) |
| ydcT | b1441 | 7.709 | 2.537 | 1.977 | putative sperimidine/putrescine transport protein (ABC superfamily, atp_bind) |
| ydcU | b1442 | 2.812 | 1.141 | 1.151 | putative sperimidine/putrescine transport protein (ABC superfamily, membrane) |
| ydcV | b1443 | 11.682 | 0.000 | 2.352 | putative sperimidine/putrescine transport protein (ABC superfamily, membrane) |
| ydcW | b1444 | 7.009 | 1.711 | 1.897 | putative aldehyde dehydrogenase |
| yddV | b1490 | 2.008 | 3.327 | 1.394 | conserved protein |
| yddX | b1481 | 2.726 | 4.638 | 4.102 | unknown CDS |
| ydeI | b1536 | 0.000 | 26.424 | 6.383 | conserved hypothetical protein |
| ydfO | b1549 | 1.601 | 2.188 | 1.294 | Qin prophage |
| ydfX | b1568 | 1.793 | 3.095 | 0.000 | Qin prophage |
| ydgA | b1614 | 2.992 | 5.893 | 2.632 | conserved protein |
| ydgE | b1599 | 1.466 | 2.094 | 1.249 | multidrug transport protein (SMR superfamily) |
| ydhF | b1647 | 1.358 | 3.738 | 1.957 | putative oxidoreductase, NAD(P)-linked domain |
| ydhJ | b1644 | 2.210 | 4.285 | 2.215 | putative multidrug resistance membrane protein |
| ydhK | b1645 | 1.502 | 2.146 | 1.275 | putative enzyme with PLP-dependent transferase domain |
| ydhL | b1648 | 0.000 | 4.382 | 2.495 | unknown CDS |
| ydhM | b1649 | 1.652 | 2.183 | 1.342 | putative transcriptional regulator with homeodomain-like DNA binding domain (TetR/AcrR family) |
| ydiN | b1691 | 2.319 | 1.225 | 0.965 | putative transport protein (MFS family) |
| ydiV | b1707 | 0.000 | 1.037 | 3.471 | conserved protein |
| ydiZ | b1724 | 4.890 | 6.105 | 1.879 | conserved hypothetical protein |
| ydjL | b1776 | 2.201 | 1.098 | 1.081 | putative dehydrogenase with NAD(P)-binding and GroES domains |
| ydjN | b1729 | 2.628 | 0.691 | 0.181 | putative transport protein |
| ydjZ | b1752 | 2.196 | 3.805 | 1.536 | putative membrane protein |
| yeaG | b1783 | 19.439 | 26.607 | 21.478 | conserved protein, nucleotide triphosphate hydrolase domain |
| yeaH | b1784 | 11.885 | 14.655 | 14.734 | conserved hypothetical protein |
| yeaI | b1785 | 1.234 | 2.797 | 1.169 | putative membrane protein |
| yeaQ | b1795 | 0.000 | 8.730 | 4.178 | conserved hypothetical protein |
| yeaW | b1802 | 1.900 | 3.975 | 4.246 | putative di(mono)oxygenase, alpha subunit with ISP and Bet v1-like domains |
| yebF | b1847 | 4.036 | 2.788 | 2.171 | conserved hypothetical protein |
| yebS | b1833 | 2.077 | 2.502 | 2.479 | putative membrane protein |
| yebT | b1834 | 2.120 | 2.644 | 1.795 | putative membrane protein |
| yebW | b1837 | 2.231 | 3.750 | 1.942 | unknown CDS |
| yedK | b1931 | 1.901 | 3.656 | 3.143 | conserved hypothetical protein |
| yedP | b1955 | 1.408 | 2.618 | 2.012 | conserved protein, phosphatase-like domain |
| yedR | b1963 | 1.332 | 2.337 | 1.510 | unknown CDS |
| yedU | b1967 | 6.016 | 15.643 | 4.887 | molecular chaperone independent of ATP/ADP cycle, with Class I glutamine amidotransferase-like domain |
| yedY | b1971 | 3.105 | 3.446 | 1.822 | putative reductase with Sulfite oxidase, middle catalytic domain |
| yeeF | b2014 | 0.000 | 2.185 | 1.084 | putative amino acid transport protein (APC family) |
| yeeJ | b1978 | 2.348 | 2.109 | 1.483 | putative transcriptional regulator |
| yeeP | b1999 | 2.799 | 4.868 | 3.081 | CP4-44 prophage; putative GTP-binding factor |
| yegP | b2080 | 17.851 | 17.674 | 7.858 | conserved hypothetical protein |
| yegS | b2086 | 8.865 | 6.521 | 13.932 | conserved protein with PDZ-like domain |
| yehE | b2112 | 0.000 | 8.156 | 5.841 | conserved hypothetical protein |
| yehX | b2129 | 1.945 | 7.563 | 1.810 | putative glycine betaine/choline transport protein with CBS regulatory domain (ABC superfamily, atp_bind) |
| yehY | b2130 | 2.097 | 2.853 | 1.453 | putative glycine betaine/choline transport protein, osmoprotection (ABC superfamily, membrane) |
| yehZ | b2131 | 5.606 | 14.355 | 2.366 | putative glycine betaine/choline transport protein, osmoprotection (ABC superfamily, peri_bind) |
| yfbF | b2254 | 2.017 | 3.008 | 1.083 | putative sugar transferase with nucleotide-diphospho-sugar transferase domain |
| yfcF | b2301 | 2.545 | 2.438 | 2.403 | putative glutathione S-transferase enzyme with thioredoxin-like domain |
| yfhB | b2560 | 1.666 | 2.151 | 1.083 | conserved protein, with a phosphatase-like domain |
| yfiH | b2593 | 1.363 | 2.040 | 1.210 | conserved hypothetical protein |
| yfiL | b2602 | 3.219 | 3.625 | 3.745 | conserved hypothetical protein |
| ygaE | b2664 | 4.842 | 10.085 | 4.299 | putative transcriptional repressor with DNA-binding Winged helix domain (GntR familiy) |
| ygaF | b2660 | 5.284 | 6.566 | 5.176 | putative enzyme |
| ygaM | b2672 | 9.390 | 14.125 | 13.022 | conserved hypothetical protein |
| ygaU | b2665 | 7.539 | 11.749 | 8.505 | conserved hypothetical protein with LysM domain |
| ygbA | b2732 | 2.665 | 2.801 | 2.083 | conserved hypothetical protein |
| ygcF | b2777 | 1.395 | 2.580 | 1.483 | putative coenzyme PQQ synthesis protein with nitrogenase iron-molybdenum domain |
| ygcJ | b2758 | 1.740 | 2.184 | 1.239 | conserved hypothetical protein |
| ygcO | b2767 | 1.176 | 2.442 | 1.542 | putative 4Fe-4S ferredoxin-type protein |
| ygdI | b2809 | 2.621 | 3.758 | 3.081 | unknown CDS |
| ygfJ | b2877 | 1.696 | 3.978 | 1.626 | conserved protein with nucleotide-diphospho-sugar transferase domain |
| ygfS | b2886 | 3.058 | 5.916 | 3.656 | putative 4Fe-4S ferredoxin-type protein |
| yggE | b2922 | 3.347 | 2.572 | 2.523 | conserved protein |
| yghA | b3003 | 4.546 | 26.142 | 6.541 | putative oxidoreductase, NAD(P)-binding domain |
| yghF | b2970 | 3.887 | 2.310 | 1.822 | putative protein exporter (General Secretory Pathway) |
| ygiL | b3043 | 2.884 | 5.483 | 0.942 | putative fimbrial-like adhesin protein |
| ygiW | b3024 | 2.855 | 2.673 | 2.159 | conserved hypothetical protein |
| ygjG | b3073 | 8.926 | 16.904 | 7.447 | putrescine:2-oxoglutaric acid aminotransferase, PLP-dependent |
| yhaO | b3110 | 0.000 | 3.532 | 0.000 | putative transport protein (HAAAP family) |
| yhbO | b3153 | 4.936 | 22.997 | 12.097 | putative intracellular proteinase with Class I glutamine amidotransferase-like domain |
| yhbP | b3154 | 1.068 | 2.001 | 1.360 | conserved protein |
| yhbW | b3160 | 2.309 | 5.070 | 1.042 | putative monooxygenase with luciferase-like ATPase activity |
| yhcH | b3221 | 1.533 | 3.032 | 1.008 | conserved hypothetical protein |
| yhcN | b3238 | 1.521 | 3.441 | 1.093 | conserved hypothetical protein |
| yhdH | b3253 | 1.244 | 2.929 | 1.451 | putative dehydrogenase, NAD(P)-binding domain and GroES-like domain |
| yhfG | b3362 | 6.030 | 8.401 | 5.947 | conserved hypothetical protein |
| yhfN | b3371 | 2.348 | 3.067 | 1.128 | fructoselysine-6-P deglycase |
| yhfW | b3380 | 4.003 | 6.668 | 4.460 | putative phosphopentomutase. alkaline phosphatase-like domain |
| yhhA | b3448 | 4.052 | 8.305 | 3.281 | conserved protein |
| yhhT | b3474 | 2.303 | 3.887 | 2.917 | putative permease (PerM family) |
| yhiM | b3491 | 7.762 | 2.489 | 2.371 | putative transport protein |
| yhiP | b3496 | 1.109 | 4.838 | 1.017 | putative peptide transport protein (POT family) |
| yhiU | b3513 | 3.483 | 1.378 | 0.985 | multidrug resistance protein (lipoprotein) |
| yhjD | b3522 | 4.105 | 5.929 | 7.239 | putative membrane protein |
| yhjG | b3524 | 3.887 | 7.107 | 4.581 | conserved protein |
| yhjQ | b3534 | 1.701 | 2.485 | 1.687 | putative cell division protein, nucleotide triphosphate hydrolase domain |
| yhjS | b3536 | 1.892 | 2.379 | 1.294 | conserved hypothetical protein |
| yhjY | b3548 | 3.268 | 4.402 | 1.986 | putative lipase |
| yiaC | b3550 | 4.052 | 2.992 | 1.209 | putative acyl-CoA N-acyltransferase |
| yiaF | b3554 | 3.929 | 3.383 | 1.302 | conserved protein |
| yiaG | b3555 | 8.401 | 9.204 | 4.080 | putative transcriptional regulator with DNA-binding domain |
| yiaH | b3561 | 1.533 | 2.377 | 2.533 | putative membrane protein |
| yibF | b3592 | 1.496 | 2.600 | 1.032 | putative glutathione S-transferase enzyme with thioredoxin-like domain |
| yibG | b3596 | 2.401 | 1.891 | 1.127 | conserved hypothetical protein |
| yibI | b3598 | 3.368 | 3.422 | 1.561 | unknown CDS |
| yidS | b3690 | 2.021 | 2.578 | 1.383 | putative oxidoreductase with FAD/NAD(P)-binding domain |
| yieK | b3718 | 1.709 | 2.679 | 1.822 | putative hexosamine-P deaminase/isomerase |
| yieM | b3745 | 1.672 | 2.017 | 1.256 | conserved protein with Integrin A (or I) domain |
| yieN | b3746 | 2.222 | 2.696 | 1.672 | multimodular: putative transcriptional regulator (N-terminal); unknown function (C-terminal) |
| yihI | b3866 | 1.510 | 2.419 | 1.652 | conserved protein |
| yiiM | b3910 | 1.725 | 2.080 | 1.738 | conserved hypothetical protein |
| yjaD | b3996 | 1.212 | 2.412 | 2.017 | conserved hypothetical protein, MutT-like protein |
| yjbB | b4020 | 1.809 | 2.145 | 1.311 | putative transport protein (PNaS family)/regulator/enzyme |
| yjbJ | b4045 | 21.962 | 20.091 | 7.597 | unknown CDS with YmbJ domain |
| yjcS | b4083 | 10.732 | 10.375 | 4.710 | putative enzyme with 2 metallo-hydrolase/oxidoreductase domains, 2 TPR-like domains, sterol carrier protein domain |
| yjdC | b4135 | 2.491 | 2.342 | 2.457 | putative regulator with homeodomain-like DNA binding domain with homeodomain-like DNA binding domain (TetR/AcrR family) |
| yjdI | b4126 | 8.414 | 9.780 | 7.145 | conserved hypothetical protein |
| yjdJ | b4127 | 6.339 | 9.340 | 6.961 | putative acyl-CoA N-acyltransferase |
| yjeB | b4178 | 2.223 | 2.487 | 2.075 | putative regulator with winged helix domain |
| yjeE | b4168 | 1.770 | 2.551 | 1.304 | putative enzyme with nucleoside triP hydrolase domain |
| yjeF | b4167 | 3.516 | 4.887 | 1.956 | putative kinase with ribokinase-like domain |
| yjeS | b4166 | 3.192 | 4.450 | 1.845 | conserved protein with 4Fe-4S ferredoxin-type domain |
| yjfN | b4188 | 1.391 | 2.417 | 2.508 | conserved hypothetical protein |
| yjfO | b4189 | 1.064 | 4.875 | 2.608 | conserved protein |
| yjfY | b4199 | 3.664 | 8.617 | 5.188 | conserved hypothetical protein |
| yjgA | b4234 | 2.089 | 1.671 | 2.312 | putative transport protein (ABC superfamily, atp_bind) |
| yjgB | b4269 | 6.977 | 10.798 | 7.402 | putative alcohol dehydrogenase with NAD(P)-binding and GroES domains |
| yjgG | b4247 | 3.617 | 3.867 | 2.216 | unknown CDS |
| yjgH | b4248 | 6.044 | 4.695 | 3.057 | putative translation factor |
| yjgR | b4263 | 2.698 | 2.938 | 2.735 | putative enzyme contains nucleoside triP hydrolase domain |
| yjhS | b4309 | 1.995 | 4.512 | 0.000 | conserved protein |
| yjhT | b4310 | 3.053 | 7.042 | 3.048 | putative enzyme contains galactose oxidase-like central domain |
| yjiD | b4326 | 1.259 | 2.061 | 1.107 | unknown CDS |
| yjiN | b4336 | 0.000 | 6.847 | 3.447 | putative transmembrane protein |
| yjiV | b4343 | 1.119 | 2.183 | 1.307 | conserved hypothetical protein |
| yjjN | b4358 | 0.882 | 2.146 | 0.908 | putative dehydrogenase with NAD(P)-binding and GroES domains |
| yjjU | b4377 | 2.688 | 1.347 | 0.781 | putative transcriptional regulator |
| yjjV | b4378 | 2.342 | 1.136 | 0.756 | putative hydrolase with metallo-dependent hydrolase domain |
| yjjY | b4402 | 2.357 | 2.592 | 1.404 | unknown CDS |
| ykfB | b0250 | 1.152 | 2.053 | 1.293 | CP4-6 prophage |
| yliI | b0837 | 4.710 | 7.931 | 4.130 | putative dehydrogenase with soluble quinoprotein glucose dehydrogenase domain |
| ymdC | b1046 | 1.687 | 2.346 | 1.767 | putative synthase with phospholipase D/nuclease domain |
| ymgA | b1165 | 4.977 | 5.866 | 4.631 | unknown CDS |
| ymgB | b1166 | 3.304 | 3.840 | 0.000 | unknown CDS |
| ymgE | b1195 | 0.000 | 8.676 | 5.861 | conserved hypothetical protein |
| yncB | b1449 | 2.075 | 9.297 | 2.698 | putative dehydrogenase, with NAD(P)-binding and GroES-like domains |
| yncC | b1450 | 2.305 | 2.979 | 2.080 | putative transcriptional regulator with DNA-binding Winged helix domain (GntR familiy) |
| yncD | b1451 | 1.327 | 3.670 | 0.882 | putative outer membrane porin protein |
| yneB | b1517 | 1.212 | 4.868 | 2.094 | putative aldolase with ribulose-phoshate binding barrel |
| ynfC | b1585 | 1.694 | 2.829 | 1.238 | conserved hypothetical protein |
| ynfD | b1586 | 2.615 | 4.332 | 4.064 | conserved hypothetical protein |
| ynhG | b1678 | 5.462 | 8.643 | 6.714 | putative ATP synthase subunit with LysM domain |
| yniA | b1725 | 1.030 | 4.372 | 1.631 | conserved protein with protein kinase-like domain |
| ynjA | b1753 | 1.013 | 3.136 | 1.575 | conserved hypothetical protein with antioxidant defence protein AhpD domain |
| ynjF | b1758 | 2.949 | 2.866 | 2.687 | putative transferase |
| yodA | b1973 | 0.000 | 2.851 | 0.000 | metal-binding protein, cadmium-induced |
| yodD | b1953 | 8.492 | 18.197 | 15.159 | unknown CDS |
| yohC | b2135 | 2.789 | 3.167 | 0.000 | putative transport protein |
| yohD | b2136 | 2.508 | 3.975 | 1.471 | putative integral membrane protein |
| yohF | b2137 | 3.972 | 11.263 | 5.212 | putative oxidoreductase with NAD(P)-binding domains |
| yojI | b2211 | 3.015 | 0.947 | 1.056 | multimodular: putative transport proteins (ABC superfamily, membrane (N-terminal), atp_bind (C-terminal)) |
| yphA | b2543 | 2.348 | 3.383 | 2.533 | putative transmembrane protein |
| yphG | b2549 | 2.639 | 2.510 | 1.538 | putative transposase with tetratricopeptide repeats (TPR) domain |
| yqaE | b2666 | 2.415 | 4.783 | 3.162 | putative transport protein (YqaE family) |
| yqeB | b2875 | 0.812 | 2.017 | 1.025 | conserved protein, NAD(P)-binding domain |
| yqjC | b3097 | 6.771 | 14.060 | 5.168 | conserved protein |
| yqjD | b3098 | 4.879 | 4.750 | 4.585 | conserved hypothetical protein |
| yqjE | b3099 | 5.264 | 5.358 | 4.652 | conserved protein |
| yqjF | b3101 | 1.180 | 4.305 | 1.642 | putative membrane protein |
| yqjG | b3102 | 4.768 | 9.030 | 5.649 | putative glutathione S-transferase enzyme with thioredoxin-like domain |
| yqjK | b3100 | 3.428 | 5.602 | 3.799 | conserved hypothetical protein |
| yraR | b3152 | 1.624 | 2.251 | 1.780 | putative NADH dehydrogenase with NAD(P)-binding domain |
| yrhB | b3446 | 1.959 | 2.145 | 0.000 | unknown CDS |
| ysgA | b3830 | 2.689 | 2.265 | 3.684 | putative dienelactone hydrolase |
| ytfI | b4215 | 1.047 | 2.574 | 1.782 | conserved hypothetical protein |
| ytfJ | b4216 | 1.105 | 3.268 | 1.364 | conserved hypothetical protein |
| ytfK | b4217 | 2.178 | 4.550 | 2.301 | unknown CDS |
| zitB | b0752 | 1.586 | 3.162 | 1.066 | Zn(II) transport protein (CDF family) |
| zupT | b3040 | 2.390 | 3.141 | 1.725 | Zn transport protein (ZIP family) |