Quantitative Biology in Space and Time

Molecular Cell Biology
Head: Prof. Ann Ehrenhofer-Murray
Our research interests are in the fields of epigenetics/ epigenomics and epitranscriptomics. We investigate how cells organize their DNA in chromatin in the nucleus, and how modifications on histones and histone variants affect gene expression, silencing and chromosome segregation. Furthermore, we study how tRNA modifications regulate protein translation.

Plant Physiology
Head: N.N. komm. Leitung Prof. Christian Schmitz-Linneweber
We are interested in the unique properties and functions of plant cells, such as the primary metabolism, pigment biosynthesis and photosynthesis in plastids and focus our efforts on exploring the metabolic control of tetrapyrrole biosynthesis.

Optobiology
Head: Prof. Marina Mikhaylova
Our lab’s overall aim our lab is to understand what defines dendritic compartments as "plasticity units". Our research questions encompass plasticity and stability of individual synapses, synaptic diversity and communication between nearby synapses. The role of the microtubule and actin cytoskeleton; trafficking rules controlling organelle transport and positioning are particularly interesting.


Structural Biology and Biochemistry
Head: Prof. Holger Dobbek
We apply protein crystallography and bioinformatics, together with biochemical and biophysical methods to study energy transforming reactions in Nature. Short-term goals are a better understanding of the enzymes involved and their evolution. The long-term goal is to develop (bio)catalysts for the energy-efficient use of CO2 and CO and the (bio)degradation of pollutants.

Plant Cell and Molecular Biology
Head: Prof. Kerstin Kaufmann
Our research is centered on the question, how cell and organ differentiation is encoded in the genome, and how this code is read and executed - depending on growth stage of the plant and on environmental factors.
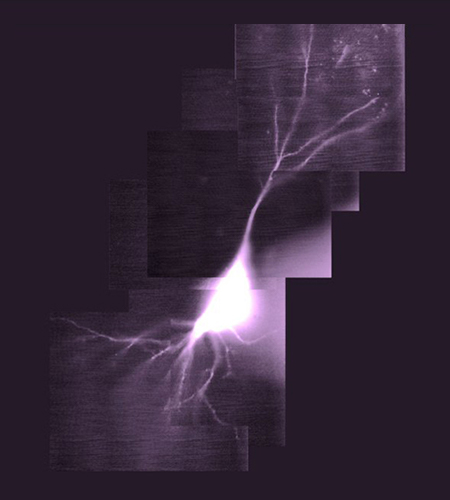

Cellular Biophysics
Head: Prof. Andrew Plested
In the Cellular Biophysics group, we study glutamate receptors and other components of fast synaptic transmission.
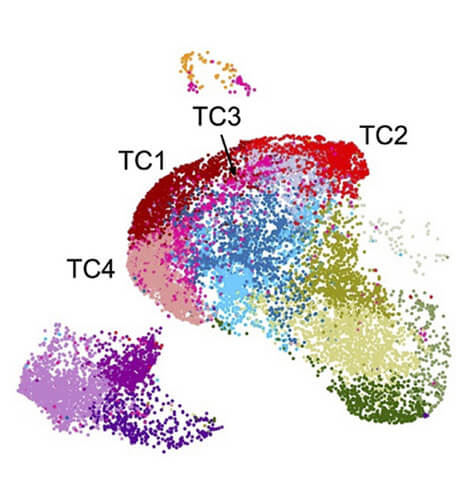

RNA Biology and Posttranscriptional Regulation
Head: Prof. Markus Landthaler
The Landthaler lab is interested in system-wide understanding of posttranscriptional regulation in mammalian cells in response to changes in their immediate environment.
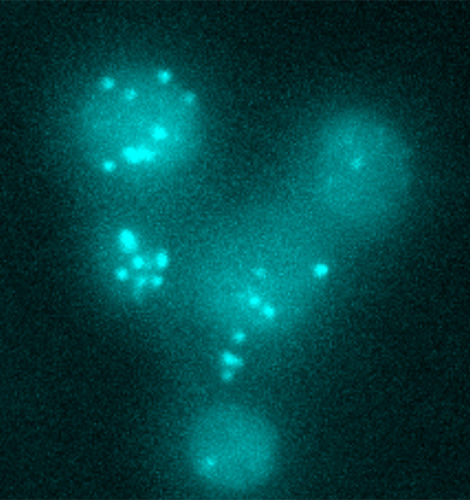

Theoretical Biophysics
Head: Prof. Edda Klipp
We study complex biological phenomena by combining computational approaches with experimental methods. Our research interests range from modeling specific signaling and metabolic pathways to whole-cell modeling using e.g. ODE, Boolean and agent-based modeling techniques.
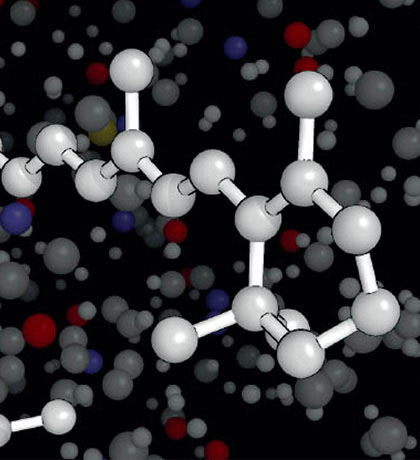

Experimental Biophysics
Head: Prof. Peter Hegemann
Our research group studies the structure-function relation of sensory photoreceptors from algae, fungi and bacteria.
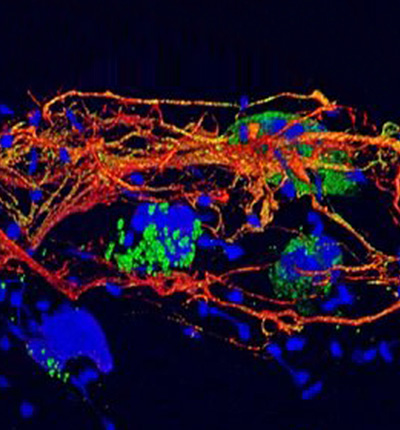

Cellular Microbiology
Head: Prof. Arturo Zychlinsky
Chromatin evolved to have an immune function in eukaryotes. Neutrophils, the most abundant white blood cell in humans, expel modified chromatin, called Neutrophil Extracellular Traps (NETs) to fight infections and regulate the immune system. Our department is taking various approaches to figure out the mechanism of NET formation.
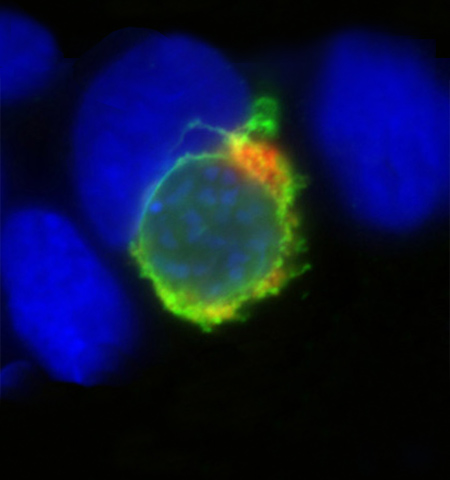

Molecular parasitology
Head: Prof. Kai Matuschewski
Eukaryotic pathogens, e.g. protozoa and helminths, are integral parts of ecosystems, and >50% of all recent animals adopted a parasitic life style. A molecular understanding of the mechanisms that drive arthropod-borne transmission, parasite stage conversion, and immune evasion are central for innovative evidence-based strategies for drug and vaccine development.
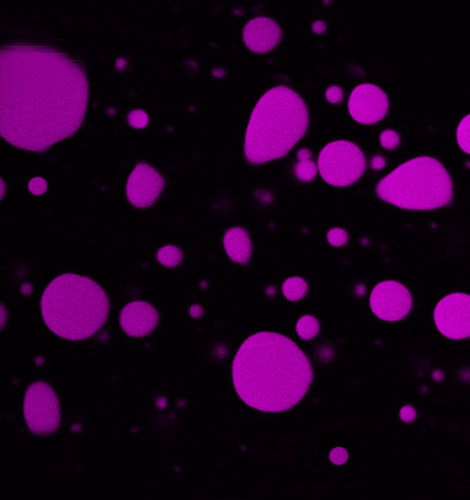

Molecular Genetics
Head: Prof. Christian Schmitz-Linneweber
We are investigating the genetic and molecular basis of nuclear-organellar interactions in plants and apicomplexan parasites. Specifically, we study novel eukaryotic RNA binding proteins used by the cell to manipulate chloroplast and mitochondrial RNAs - essential processes for setting up the respiratory chain and the photosynthetic machinery.


Regulation in Infection Biology
Head: Prof. Emmanuelle Charpentier
We study fundamental regulatory mechanisms in the pathogenic bacterium Streptococcus pyogenes (the source of the CRISPR-Cas9 gene editing technology). We use an interdisciplinary approach combining cutting-edge methodologies—bioinformatics, omics, genetics, molecular biology, structural biology, biochemistry, physiology, and cell infection—to identify new molecules and mechanisms and decipher their origins, functions, and modes of action at the molecular and cellular levels.
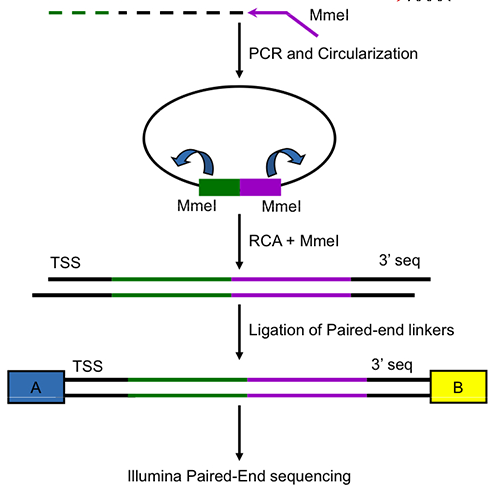

Computational regulatory genomics
Head: Prof. Uwe Ohler
Our lab develops and applies genomics and computational approaches to understand mechanisms of gene regulation in eukaryotic organisms. Our long term goal is to investigate how regulatory networks enable the correct development of complex organisms, with their multitude of cell types that carry out different functions despite the same genome.


Molecular Parasitology / Canberra
Head: Prof. Alexander Maier
The Maier lab focussed on the identification of molecules involved in malaria pathogenesis and transmission. In order to elucidate the functions of these molecules we use biochemical, molecular and cell biological techniques. The long-term goal is not only to identify and develop new intervention strategies against malaria, but also to understand the underlying biological principles.


Comparative Zoology
Head: Prof. John Nyakatura
Our team focusses on the functional morphology and evolution of land-living vertebrates. Collection-based approaches, experimental approaches and field work are integrated to study form-function relationships on an organismic level in the context of evolution.
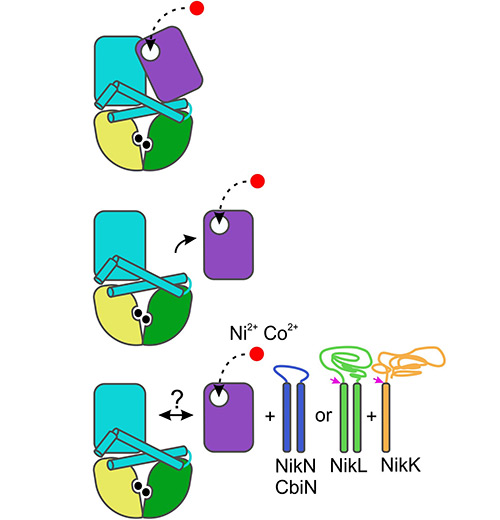

Microbiology
Head: Prof. Thomas Eitinger
A research focus is on energy-coupling factor (ECF) transporters, a widespread, mechanistically distinct and underexplored group of ATP-binding cassette transporters in bacteria and archaea (and perhaps in plants). Among prokaryotes, these uptake systems mediate high-affinity uptake of water-soluble vitamins and transition metal ions into the cells. In organisms with restricted biosynthetic capacity, e.g. pathogenic bacteria, ECF-type vitamin transporters are essential for viability.
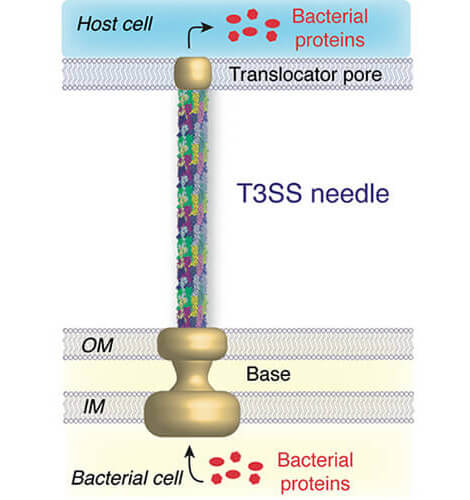

Structure and Dynamics of Biomolecules
Head: Prof. Adam Lange
We use solid-state NMR spectroscopy and a variety of other biophysical methods to study protein structure and dynamics. The systems of interest comprise membrane proteins in the context of a native-like lipid bilayer environment and supramolecular assemblies such as type three secretion needles or cytoskeletal filaments.


Collective Information Processing
Head: Prof. Pawel Romanczuk
We investigate the interplay of self-organization and function in complex biological systems by combining mathematical modeling and analysis of experimental data. Our main research focus are mechanisms and functional benefits of collective behavior from cellular ensembles to animal groups. Further topics include modeling of stochastic motility and spatially-explicit evolutionary games.
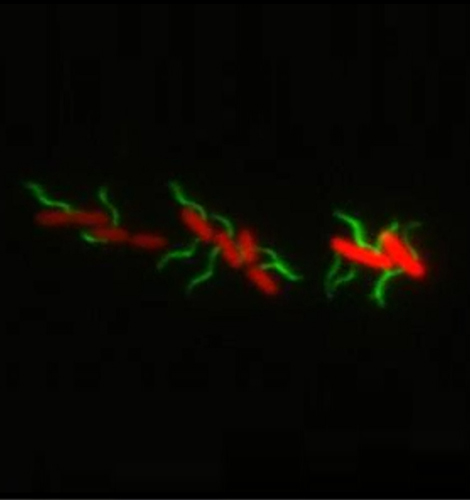

Molecular Microbiology
Head: Prof. Marc Erhardt
Bacteria use a complex macromolecular machine, the so-called flagellum, to move in liquid environments. Flagella-mediated motility is also important for the pathogenicity of many pathogens such as Salmonella. We employ genetic engineering, biochemistry and fluorescent microscopy techniques to understand the regulation, self-assembly and protein export mechanisms of this fascinating nanomachine.
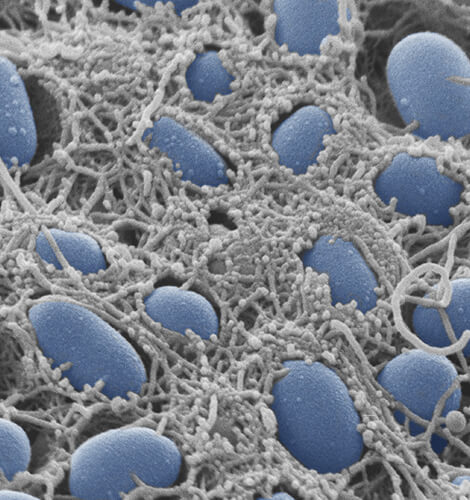

General Microbiology
Head: Prof. Regine Hengge
Research focuses on signal transduction networks and gene regulation in nutrient-limited non-growing, but highly stress-resilient bacteria and bacterial biofilms. In particular, molecular functions of second messengers such as cyclic-di-GMP in orchestrating bacterial multicellularity and its emergent properties are studied. Furthermore, we collaborate with designers, material scientists and cultural historians in interdisciplinary projects.
