Exploiting cyanobacterial Photosystem I and other Photosynthetic pigment - protein complexes for nano-photobiotechnology
Overview of photosynthesis
Oxygenic photosynthesis supplies energy to virtually all life forms on our planet. Photosynthesis is the process by which photosynthetic organisms, such as green plants, algae and cyanobacteria use sunlight to synthesize carbohydrate molecules from carbon dioxide and water. This process combines six molecules of carbon dioxide and six molecules of water to produce one molecule of glucose and six oxygen molecules. The oxygen that is produced as a byproduct of photosynthesis, is what most animals rely on, to breath and is critical to our survival (Fig. 1).
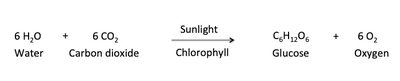
Fig. 1. Chemical equation of Photosynthesis
Photosynthetic organisms employ two large pigment-protein supercomplexes called photosystems located at the thylakoid membrane to drive the light reactions of oxygenic photosynthesis (Fig. 2). Upon excitation by sunlight, PSII catalyses the transfer of electrons from water (oxidation of water occurs at the donor side, Kok cycle) to plastoquinone (PQ, at the acceptor side). The electron leaves PSII as plastoquinol (PQH2) and traverses through the thylakoid membrane via membrane bound cytochrome b6f to the soluble electron carrier plastocyanin (PC) (or cytochrome c6 in cyanobacteria). At PSI, a light-induced electron transfer leads to the oxidation of PC and the reduction of Ferredoxin (Fd). The latter carries the electrons to FNR, where they are used for the reduction of NADP to NADPH.
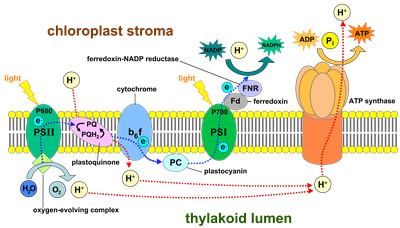
Fig. 2. Schematic view of electron transport via the PSII and PSI protein complexes along the thylakoid membrane. (Source: Wikipedia)
PS I from Thermosynechococcus (T.) elongatus
PS I from thermophilic cyanobacterium Thermosynechococcus elongatus has been crystallized and its structure has been determined to a resolution of 2.5 Å (Jordan et al. 2001). PSI can be readily isolated from T. elongatus. The trimeric PS I core complex (PSIcc) comprises three monomers forming a super complex of ca. 1 MDa molecular weight with the PsaL subunit at the interface between monomers. Each monomeric subunit consists of 12 protein subunits, 127 co-factors, including 96 chlorophylls a and 3 iron-sulphur clusters (Fx, FA and FB), performing light capture and electron transfer. Nine of the protein subunits are transmembrane and three are extrinsic subunits (PsaC, E, and D) at the stromal side (Fig.3b).
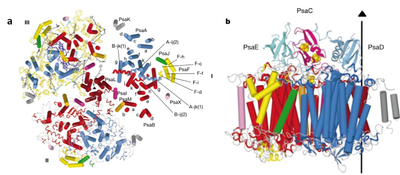
Fig.3. Structural model of PSI trimer at 2.5Å resolution. A. A view on the membrane from the stromal side (top view). B. Side view of a PSI monomer showing all proteins including the stromal subunits PsaC (pink), PsaD (turquoise), PsaE (green), and the Fe4S4 clusters. (Source: Jordan et. al., Nature, 2001)
The two core subunits PsaA and PsaB bind the majority of cofactors, including reaction center (RC) and antenna pigments. In the RC, light-induced charge separation starts at the primary electron donor P700 (a Chl a/a’ dimer), with one of the branches being more active than the other. From P700, through the primary electron acceptor A0 (a Chl a monomer), the secondary electron acceptor A1 (a phylloquinone), the electrons are transferred to the iron-sulfur cluster FX. From Fx, the electrons are finally accepted by the two iron–sulphur clusters FA and FB that are bound by subunit PsaC. The FB cluster transfers the electrons to ferredoxin.
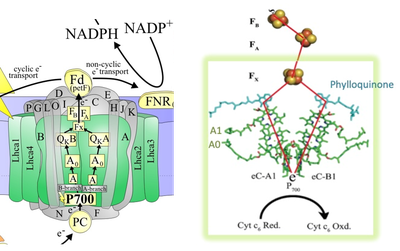
Fig. 4. Arrangement of co-factors involved in electron transfer in PSI reaction center. (Source: Molecular Mechanism of Photosynthesis, Second edition, Robert E. Blankenship (2014) and Lubner et. al. (2011))
PSI as an important model system for artificial photosynthesis
PSI from T. elongatus is exceptionally stable and retains a high rate of light-induced electron transfer under a wide range of conditions (temperature, pH, etc.) for extended period of time. The light-induced charge separation in PSI can be exploited for various applications, where the electrons are utilized to drive biomolecular transformations. Its robustness renders PSI highly suitable for biotechnological applications - such as incorporation into bio-hybrid (nano-) devices. Besides, PSI is an important model system for investigating fundamental aspect of natural and artificial photosynthesis.
Artificial photosynthesis (AP) can be utilized as a renewable source of energy, which can maintain an energy supply while minimising impact on the environment and climate. In AP, solar energy is converted with the assistance of catalytic processes and used for producing fuels and valuable products. Unlike solar photovoltaic cells, where the electricity cannot be stored, the by-products produced by AP can be stored, stockpiled and transported. However, the biggest challenge faced by AP is cost-effectiveness.
The aim of our BMBF tandem project
In cooperation with Prof. Dr. Fred Lisdat, TFH Wildau, aim of the tandem project is to develop light-activatable electrode assemblies for bio catalytic synthesis. For this purpose, as the first stage, photoactive biomolecules such as Cyt C-PSI can be combined with electrodes, so that an effective electron transfer after photo-excitation is possible. Such photobioelectrode hybrid can be established using functional structures of biological and inorganic components. In the second stage, enzyme systems will be coupled with light-activatable electrodes in order to use the generated photocurrent for the biocatalyzed synthesis of important chemical compounds. Thus, such a modular solar light-powered photobioelectrodes can be used for the production of high-value products (HVPs). The long-term goal is to increase the efficiency of conversion of light energy into biochemical recyclables.
For the first stage of this tandem project, please see the completed project and publications.
Second stage: In the current project, we plan to couple (redox-) enzymes, such as formate dehydrogenase (in co-operation with Prof. Silke Leimkühler) or Carbon monoxide dehydrogenase (in co-operation with Prof. Holger Dobbek), to photobioelectrode hybrid assembly containing Cyt C-PSI using molecular wire. In order to achieve effective electron transfer, a thiol linker (molecular wire) with a defined length will be used to link the terminal electron acceptor of PSI (PsaC) and redox enzymes. Such molecular wire can be attached by a ligand exchange mechanism where exchangeable sulfhydryl ligand is introduced to the most solvent-exposed iron atom of PsaC. A site-specific conversion of a ligating Cys residue (C14) of FB of PsaC to a Gly is required for the chemical rescuing of the cluster with a small sulfhydryl-containing molecule (Lubner et. al., 2011). PsaC protein with a mutation (C14G) will be expressed in E. coli and purified. Together with PsaD, this mutant PsaC protein will bind naturally to PSI core, whose extrinsic subunits PsaC, E and D have been previously removed using chaotropic agents. Such a mutant PsaC protein enables the binding of thiol linker to the coordination site of the FB cluster. As a result, the PSI with mutant PsaC will be provided with a linker bearing a thiol group at its free end. This enables redox enzyme to couple to the free thiol group, ultimately enabling the flow of electron from PSI to the active site of the redox enzyme.
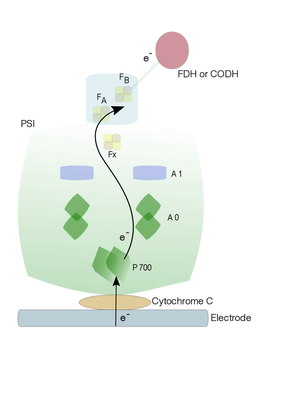
Figure 5: A schematic representation of electrode coupling with Cyt C-PSI-FDH or -CODH to generate chemical substances
Another approach can be a direct coupling of redox enzyme via molecular wires to the FX iron-sulphur cluster in PSI complex without PsaC. Chaotropic agents such as NaBr and NaI have been shown to remove external stromal subunits (Parrett et. al., 1989). Removal of external subunits (PsaC) results in the exposure of Fx and thereby reducing the distance the electron has to travel to the redox enzyme. Such a direct electron transfer from Fx to enzyme active site may improve the enzyme kinetics.
Co-operation partners:
Prof. Fred Lisdat-TH Wildau
Prof. Silke Leimkühler-University of Potsdam
Prof. Holger Dobbek-Humboldt University of Berlin
Current members working in this project:
Dr. Senthil Kumar Subramanian
Adrian Kölsch
Jelena Boyke
Literature:
Lubner et. al., PNAS (2011). Solar hydrogen-producing bionanodevice outperforms natural photosynthesis
Parrett et. al., BBA ( 1989). Purification and properties of the intact P-700 and Fx-containing Photosystem I core protein.
Jordan et. al., Nature (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution